Generalized Linear Model (GLM)
PSYC 573
GLM
Three components:
- Conditional distribution of \(Y\)
- Link function
- Linear predictor (\(\eta\))
Some Examples
| Outcome type | Support | Distributions | Link |
|---|---|---|---|
| continuous | [\(-\infty\), \(\infty\)] | Normal | Identity |
| count (fixed duration) | {0, 1, \(\ldots\)} | Poisson | Log |
| count (known # of trials) | {0, 1, \(\ldots\), \(N\)} | Binomial | Logit |
| binary | {0, 1} | Bernoulli | Logit |
| ordinal | {0, 1, \(\ldots\), \(K\)} | categorical | Logit |
| nominal | \(K\)-vector of {0, 1} | categorical | Logit |
| multinomial | \(K\)-vector of {0, 1, \(\ldots\), \(K\)} | categorical | Logit |
Mathematical Form (One Predictor)
\[ \begin{aligned} Y_i & \sim \mathrm{Dist}(\mu_i, \tau) \\ g(\mu_i) & = \eta_i \\ \eta_i & = \beta_0 + \beta_1 X_{i} \end{aligned} \]
- \(\mathrm{Dist}\): conditional distribution of \(Y \mid X\) (e.g., normal, Bernoulli, \(\ldots\))
- I.e., distribution of prediction error; not the marginal distribution of \(Y\)
- \(\mu_i\): mean parameter for the \(i\)th observation
- \(\eta_i\): linear predictor
- \(g(\cdot)\): link function
- (\(\tau\): dispersion parameter)
Illustration
Next few slides contain example GLMs, with the same predictor \(X\)
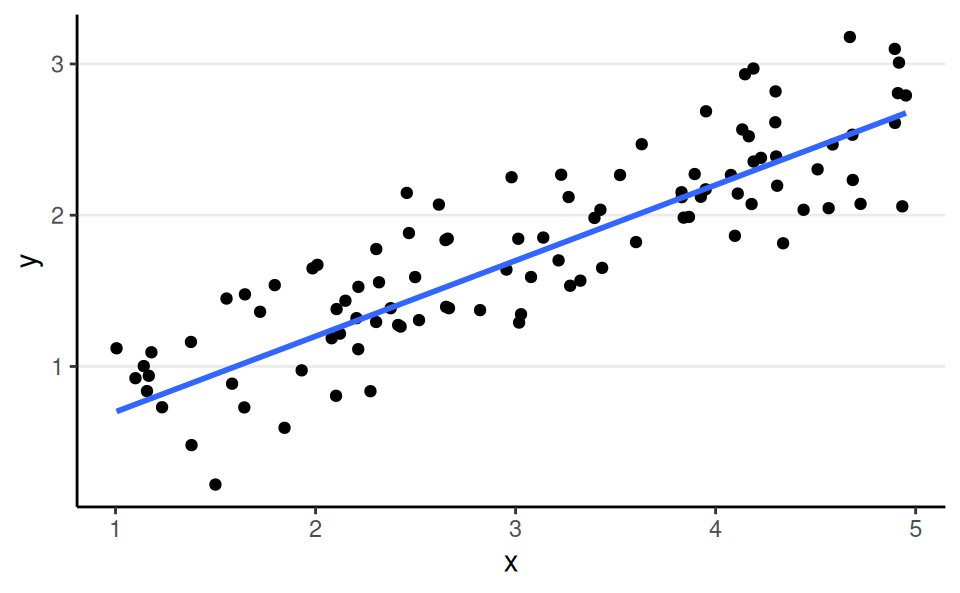
Normal, Identity Link
aka linear regression
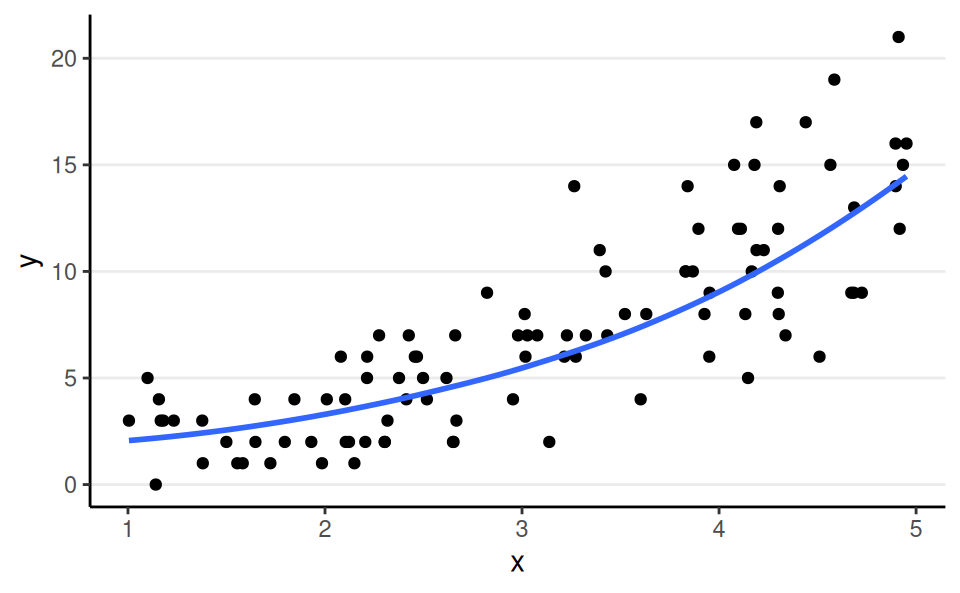
Poisson, Log Link
aka poisson regression
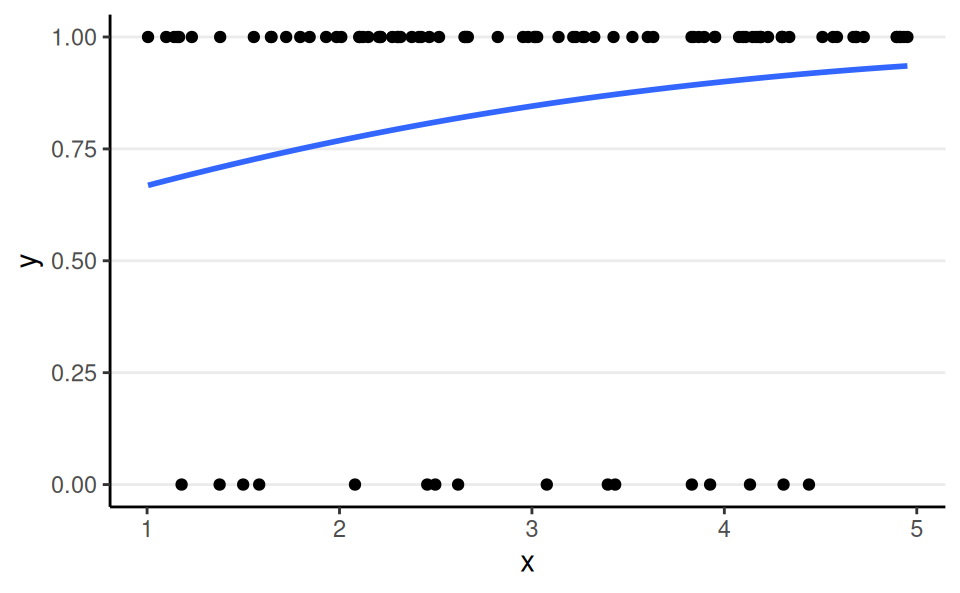
Bernoulli, Logit Link
aka binary logistic regression
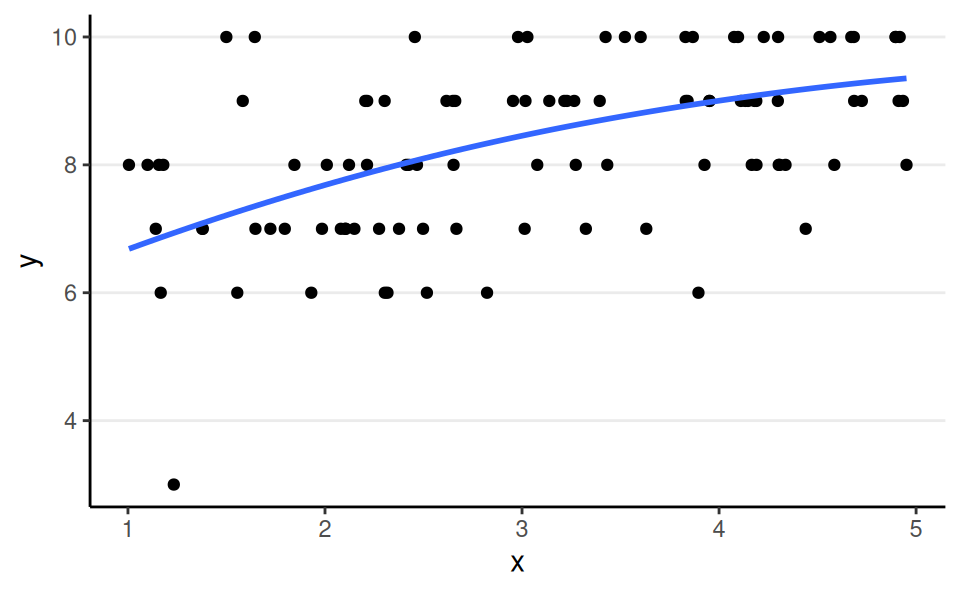
Binomial, Logit Link
aka binomial logistic regression
\[ \begin{aligned} Y_i & \sim \mathrm{Bin}(N, \mu_i) \\ \log\left(\frac{\mu_i}{1 - \mu_i}\right) & = \eta_i \\ \eta_i & = \beta_0 + \beta_1 X_{i} \end{aligned} \]
Remarks
Different link functions can be used
- E.g., identity link or probit link for Bernoulli variables
Linearity is a strong assumption
- GLM can allow \(\eta\) and \(X\) to be nonlinearly related, as long as it’s linear in the coefficients
- E.g., \(\eta_i = \beta_0 + \beta_1 \log(X_{i})\)
- E.g., \(\eta_i = \beta_0 + \beta_1 X_i + \beta_2 X_i^2\)
- But not something like \(\eta_i = \beta_0 \log(\beta_1 + x_i)\)
Logistic Regression
See exercise
Poisson Regression
count: The seizure count between two visitsTrt: Either 0 or 1 indicating if the patient received anticonvulsant therapy
\[ \begin{aligned} \text{count}_i & \sim \mathrm{Pois}(\mu_i) \\ \log(\mu_i) & = \eta_i \\ \eta_i & = \beta_0 + \beta_1 \text{Trt}_{i} \end{aligned} \]
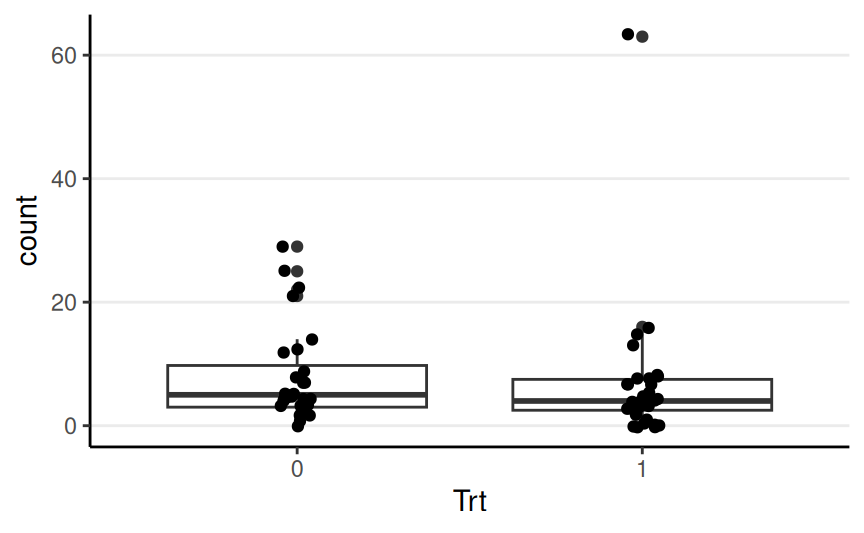
Poisson with log link
Predicted seizure rate = \(\exp(\beta_0 + \beta_1) = \exp(\beta_0) \exp(\beta_1)\) for Trt = 1; \(\exp(\beta_0)\) for Trt = 0
\(\beta_1\) = mean difference in log rate of seizure; \(\exp(\beta_1)\) = ratio in rate of seizure
Family: poisson
Links: mu = log
Formula: count ~ Trt
Data: epilepsy4 (Number of observations: 59)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept 2.07 0.07 1.94 2.21 1.00 4022 2321
Trt1 -0.17 0.10 -0.36 0.02 1.00 3435 2620
Draws were sampled using sample(hmc). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).Poisson with identity link
In this case, with one binary predictor, the link does not matter to the fit
\[ \begin{aligned} \text{count}_i & \sim \mathrm{Pois}(\mu_i) \\ \mu_i & = \eta_i \\ \eta_i & = \beta_0 + \beta_1 \text{Trt}_{i} \end{aligned} \]
\(\beta_1\) = mean difference in the rate of seizure in two weeks
| log link | identity link | |
|---|---|---|
| b_Intercept | 2.07 | 7.95 |
| [1.94, 2.21] | [6.99, 9.08] | |
| b_Trt1 | −0.17 | −1.24 |
| [−0.36, 0.02] | [−2.59, 0.07] | |
| Num.Obs. | 59 | 59 |
| R2 | 0.004 | 0.004 |
| ELPD | −345.8 | −343.8 |
| ELPD s.e. | 96.0 | 94.2 |
| LOOIC | 691.5 | 687.7 |
| LOOIC s.e. | 192.0 | 188.3 |
| WAIC | 690.2 | 689.5 |
| RMSE | 9.54 | 9.56 |