| y | s |
|---|---|
| 1 | S01 |
| 0 | S01 |
| 0 | S01 |
| 0 | S01 |
| 0 | S01 |
| 0 | S01 |
| 0 | S01 |
| 0 | S01 |
| 0 | S01 |
| 0 | S01 |
Hierarchical Models
PSYC 573
2024-09-17
Therapeutic Touch Example (N = 28)
Data Points From One Person

\(y\): whether the guess of which hand was hovered over was correct
Person S01
Binomial Model
We can use a Bernoulli model: \[ y_i \sim \mathrm{Bern}(\theta) \] for \(i = 1, \ldots, N\)
Assuming exchangeability given \(\theta\), more succint to write \[ z \sim \mathrm{Bin}(N, \theta) \] for \(z = \sum_{i = 1}^N y_i\)
- Bernoulli: Individual trial
- Binomial: total count of “1”s
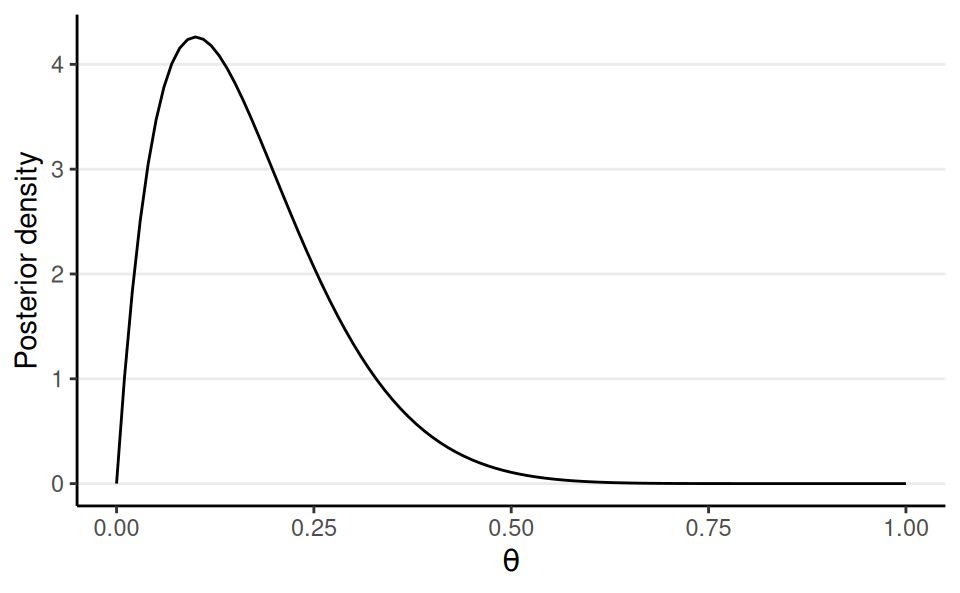
Prior: Beta(1, 1)
1 success, 9 failures
Posterior: Beta(2, 10)
Multiple People
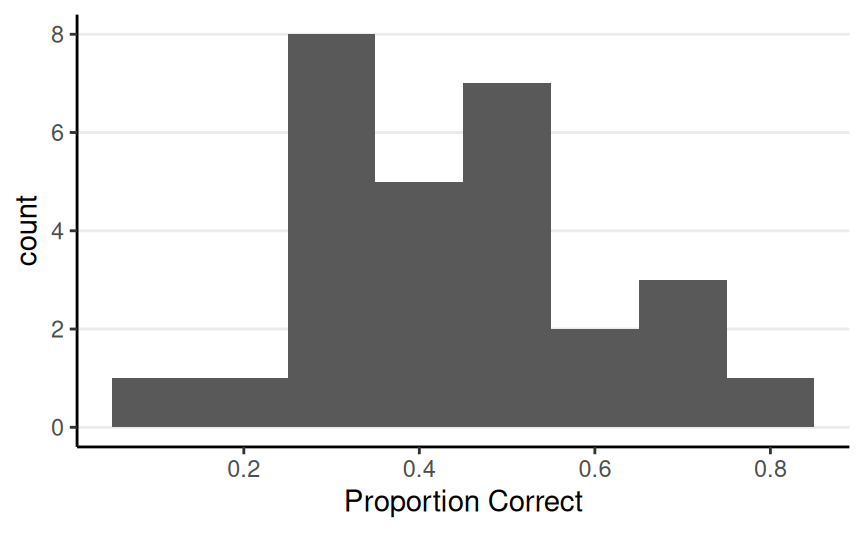
We could repeat the binomial model for each of the 28 participants, to obtain posteriors for \(\theta_1\), \(\ldots\), \(\theta_{28}\)
But . . .
Do we think our belief about \(\theta_1\) would inform our belief about \(\theta_2\), etc?
After all, human beings share 99.9% of genetic makeup
Three Positions of Pooling
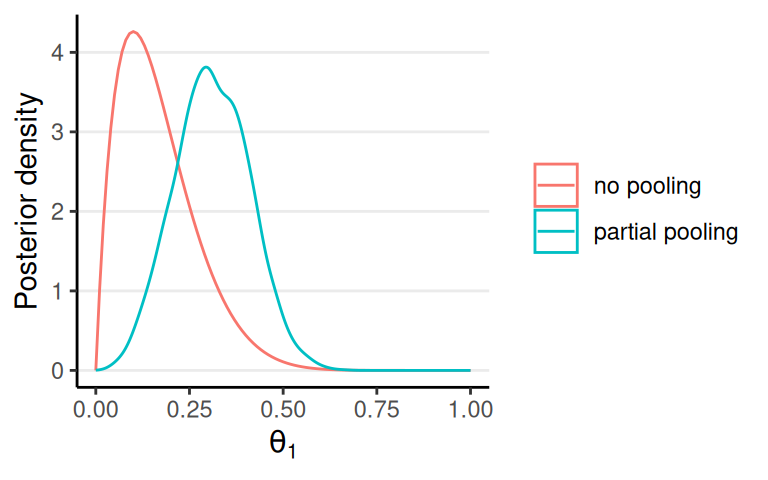
No pooling: each individual is completely different; inference of \(\theta_1\) should be independent of \(\theta_2\), etc
Complete pooling: each individual is exactly the same; just one \(\theta\) instead of 28 \(\theta_j\)’s
Partial pooling: each individual has something in common but also is somewhat different
No Pooling
Complete Pooling
Partial Pooling
Partial Pooling in Hierarchical Models
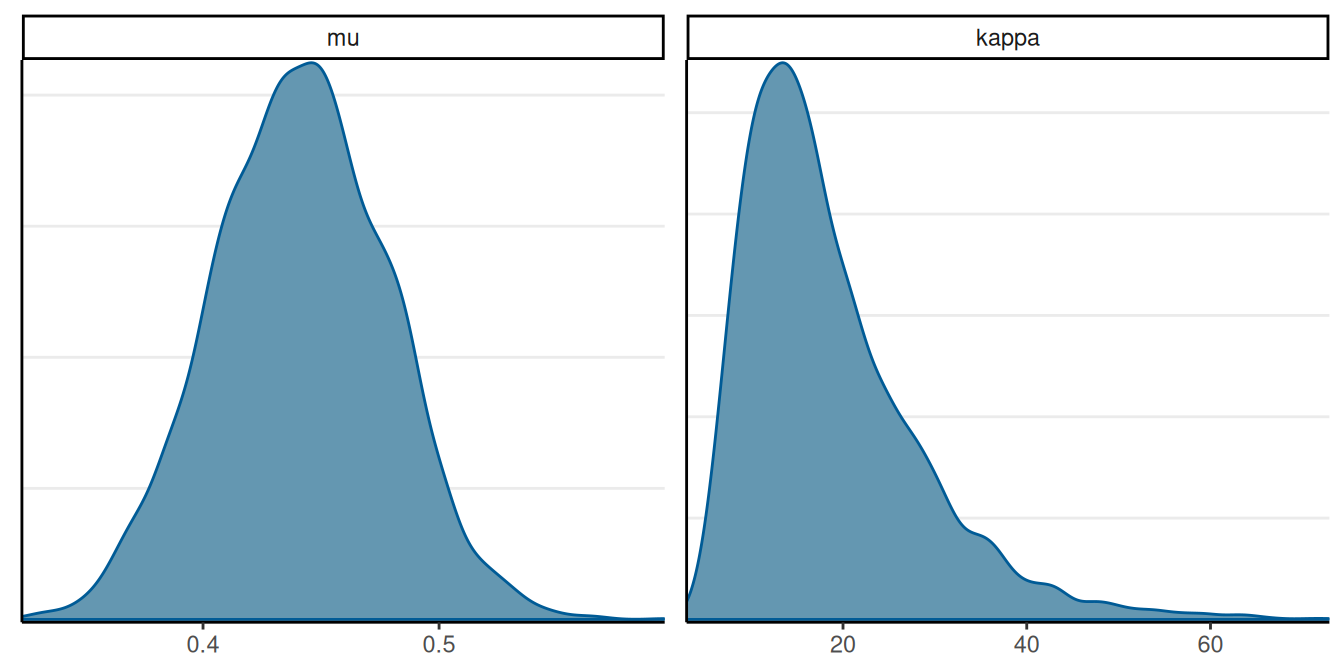
Hierarchical Priors: \(\theta_j \sim \mathrm{Beta2}(\mu, \kappa)\)
Beta2: reparameterized Beta distribution
- mean \(\mu = a / (a + b)\)
- concentration \(\kappa = a + b\)
Expresses the prior belief:
Individual \(\theta\)s follow a common Beta distribution with mean \(\mu\) and concentration \(\kappa\)
How to Choose \(\kappa\)
If \(\kappa \to \infty\): everyone is the same; no individual differences (i.e., complete pooling)
If \(\kappa = 0\): everybody is different; nothing is shared (i.e., no pooling)
We can fix a \(\kappa\) value based on our belief of how individuals are similar or different
A more Bayesian approach is to treat \(\kappa\) as an unknown, and use Bayesian inference to update our belief about \(\kappa\)
Generic prior by Kruschke (2015): \(\kappa\) \(\sim\) Gamma(0.01, 0.01)
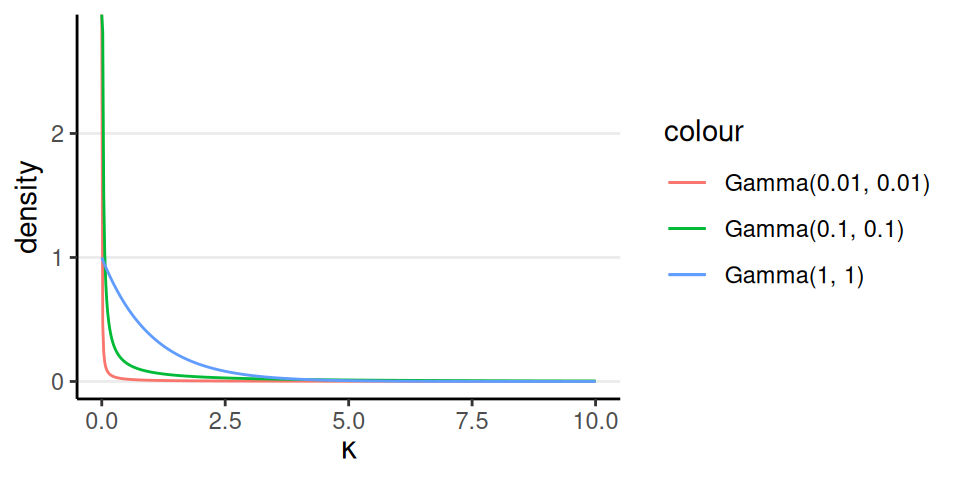
Sometimes you may want a stronger prior like Gamma(1, 1), if it is unrealistic to do no pooling
Full Model
Model: \[ \begin{aligned} z_j & \sim \mathrm{Bin}(N_j, \theta_j) \\ \theta_j & \sim \mathrm{Beta2}(\mu, \kappa) \end{aligned} \] Prior: \[ \begin{aligned} \mu & \sim \mathrm{Beta}(1.5, 1.5) \\ \kappa & \sim \mathrm{Gamma}(0.01, 0.01) \end{aligned} \]
data {
int<lower=0> J; // number of clusters (e.g., studies, persons)
array[J] int y; // number of "1"s in each cluster
array[J] int N; // sample size for each cluster
}
parameters {
// cluster-specific probabilities
vector<lower=0, upper=1>[J] theta;
real<lower=0, upper=1> mu; // overall mean probability
real<lower=0> kappa; // overall concentration
}
model {
y ~ binomial(N, theta); // each observation is binomial
// Priors
theta ~ beta_proportion(mu, kappa);
mu ~ beta(1.5, 1.5); // weak prior
kappa ~ gamma(.1, .1); // prior recommended by Kruschke
}
generated quantities {
// Prior and posterior predictive
real<lower=0, upper=1> prior_mu = beta_rng(1.5, 1.5);
real<lower=0> prior_kappa = gamma_rng(.1, .1);
vector<lower=0, upper=1>[J] prior_theta;
for (j in 1:J) {
prior_theta[j] = beta_proportion_rng(prior_mu, prior_kappa);
}
array[J] int prior_ytilde = binomial_rng(N, prior_theta);
// Posterior predictive
array[J] int ytilde = binomial_rng(N, theta);
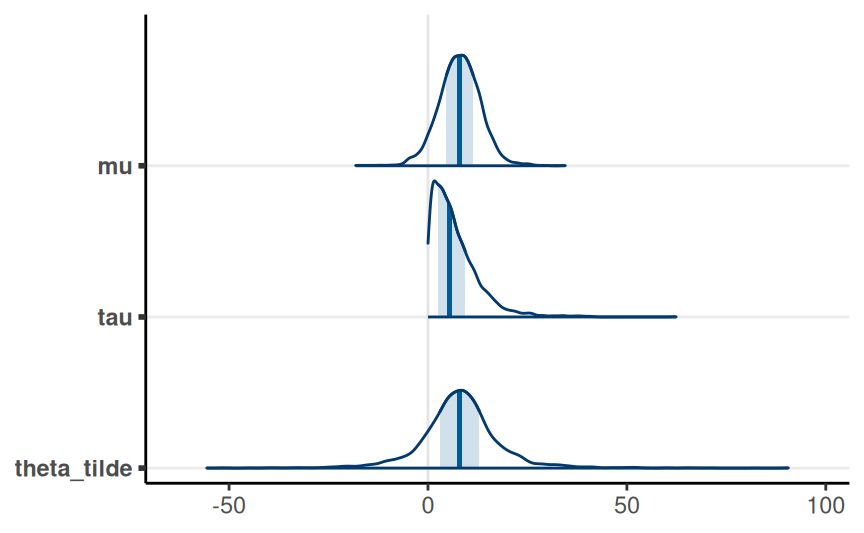
}Posterior of Hyperparameters
Shrinkage
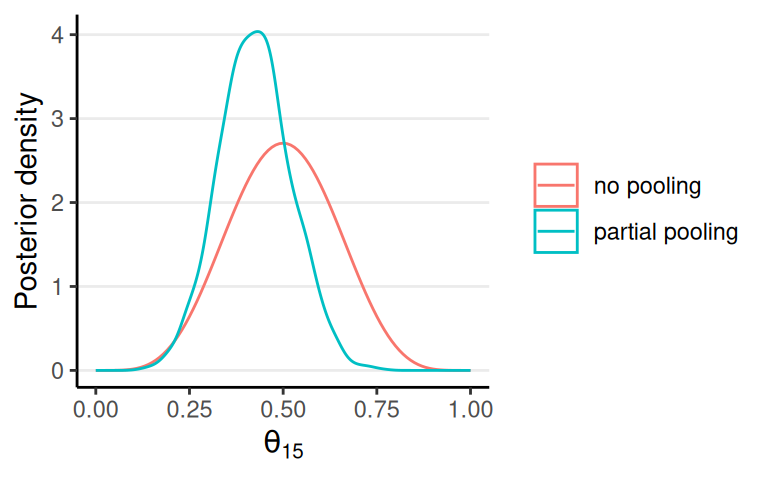
Multiple Comparisons?
Frequentist: family-wise error rate depends on the number of intended contrasts
Bayesian: only one posterior; hierarchical priors already express the possibility that groups are the same
Thus, Bayesian hierarchical model “completely solves the multiple comparisons problem.”1
Hierarchical Normal Model
Effect of coaching on SAT-V
| School | Treatment.Effect.Estimate | Standard.Error |
|---|---|---|
| A | 28 | 15 |
| B | 8 | 10 |
| C | -3 | 16 |
| D | 7 | 11 |
| E | -1 | 9 |
| F | 1 | 11 |
| G | 18 | 10 |
| H | 12 | 18 |
Model: \[ \begin{aligned} d_j & \sim N(\theta_j, s_j) \\ \theta_j & \sim N(\mu, \tau) \end{aligned} \] Prior: \[ \begin{aligned} \mu & \sim N(0, 100) \\ \tau & \sim t^+_4(0, 100) \end{aligned} \]
data {
int<lower=0> J; // number of schools
vector[J] y; // estimated treatment effects
vector<lower=0>[J] sigma; // s.e. of effect estimates
}
parameters {
real mu; // overall mean
real<lower=0> tau; // between-school SD
vector[J] eta; // standardized deviation (z score)
}
transformed parameters {
vector[J] theta;
theta = mu + tau * eta; // non-centered parameterization
}
model {
eta ~ std_normal(); // same as eta ~ normal(0, 1);
y ~ normal(theta, sigma);
// priors
mu ~ normal(0, 100);
tau ~ student_t(4, 0, 100);
}Individual-School Treatment Effects
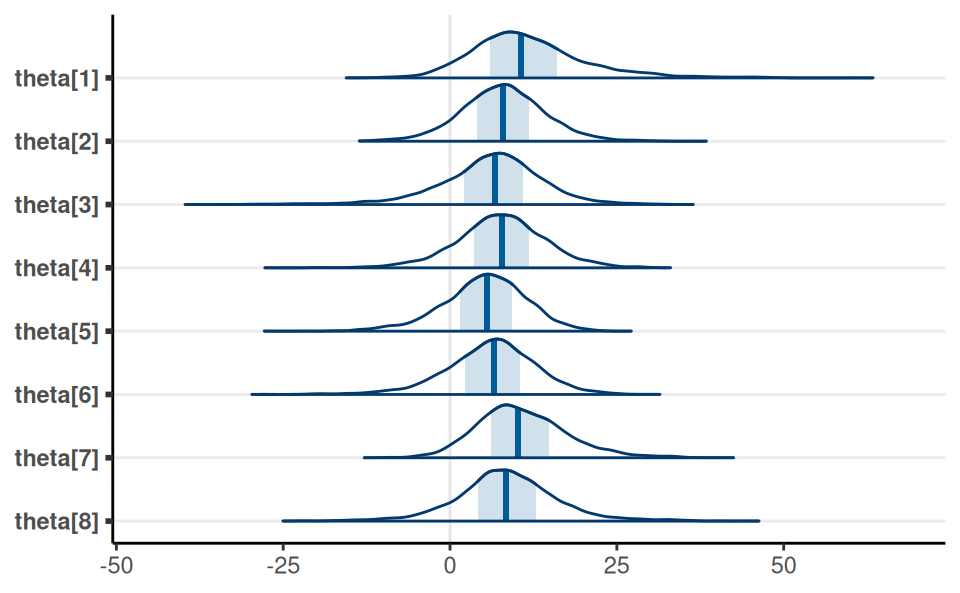
Prediction Interval
Posterior distribution of the true effect size of a new study, \(\tilde \theta\)