# Define a function for formatting numbers
comma <- function(x, d = 2) format(x, digits = d, big.mark = ",")ROC and MTMM
library(readxl) # for reading excel data
library(haven) # for reading SPSS data
library(ROCit) # for ROC analysis
library(psych)
library(tidyverse)
library(modelsummary) # for summarizing dataClassification Accuracy
Data analyzed in this paper: https://journals.sagepub.com/doi/10.1177/07342829211067128
tmp_path <- tempfile(fileext = "xlsx") # temporary file
download.file("https://osf.io/download/tsm7x/", destfile = tmp_path)
dat1 <- readxl::read_xlsx(tmp_path, na = "NA")
dat1$asd <- recode(
dat1$`Diagnosis(1=Non-typically developing; 2=ASD; 3=Neurotypical)`,
`1` = "Non-TD",
`2` = "ASD",
`3` = "TD"
)
# Filter out Neurotypical
dat1_sub <- filter(dat1, asd != "TD")First, sum the 16 items. Note that the sum is missing if any one item is missing
dat1_sub$ADEC <- rowSums(select(dat1_sub, ADEC_I01:ADEC_I16)) # NA if any item is NA
# Or treat missing as 0
dat1_sub$ADEC_rm_na <- rowSums(select(dat1_sub, ADEC_I01:ADEC_I16), na.rm = TRUE)Now, we can look at the distribution of ADEC sum score by diagnosis:
ggplot(dat1_sub, aes(x = ADEC)) +
geom_histogram() +
facet_wrap(~ asd)`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.Warning: Removed 12 rows containing non-finite values (`stat_bin()`).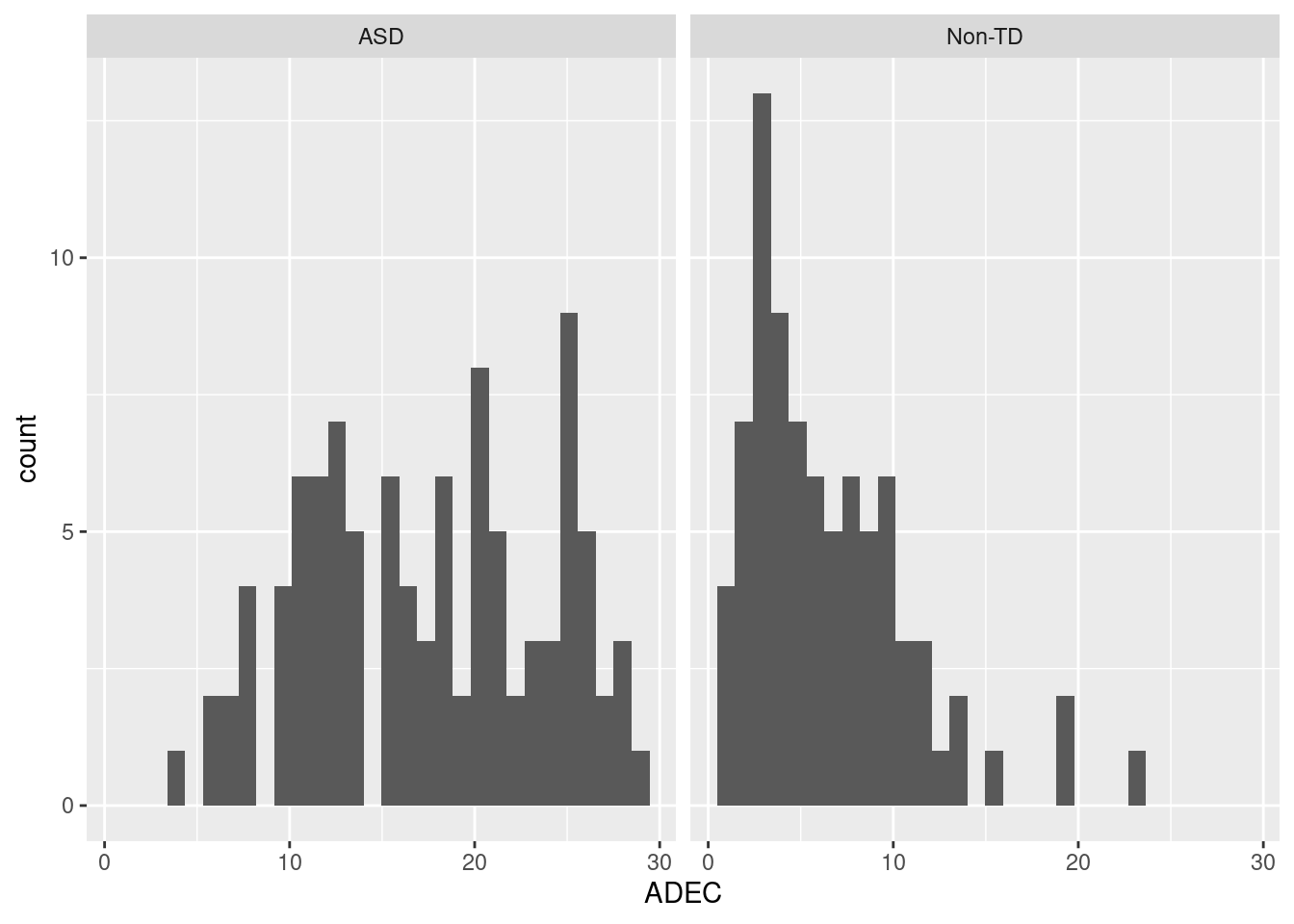
Classification Table
Let’s first use ADEC = 11 as cutoff. We then get the following contingency table
with(dat1_sub,
table(ADEC >= 11, asd)) asd
ASD Non-TD
FALSE 13 68
TRUE 86 13So the accuracy with this cutoff is (86 + 68) / 180 = 0.86. The sensitivity is 86 / (86 + 13) = 0.87, and the specificity is 68 / (68 + 13) = 0.84.
ROC
We can plot an ROC curve
compute_sens <- function(
cut, x = dat1_sub$ADEC,
crit = dat1_sub$asd == "ASD") {
tp <- sum(x >= cut & crit, na.rm = TRUE)
fn <- sum(x < cut & crit, na.rm = TRUE)
tp / (tp + fn)
}
compute_spec <- function(
cut, x = dat1_sub$ADEC,
crit = dat1_sub$asd == "ASD") {
tn <- sum(x < cut & !crit, na.rm = TRUE)
fp <- sum(x >= cut & !crit, na.rm = TRUE)
tn / (tn + fp)
}
sensitivity <- lapply(32:0, compute_sens) |>
unlist()
specificity <- lapply(32:0, compute_spec) |>
unlist()
data.frame(sensitivity, fpr = 1 - specificity) |>
ggplot(aes(x = fpr, y = sensitivity)) +
geom_point() +
geom_line()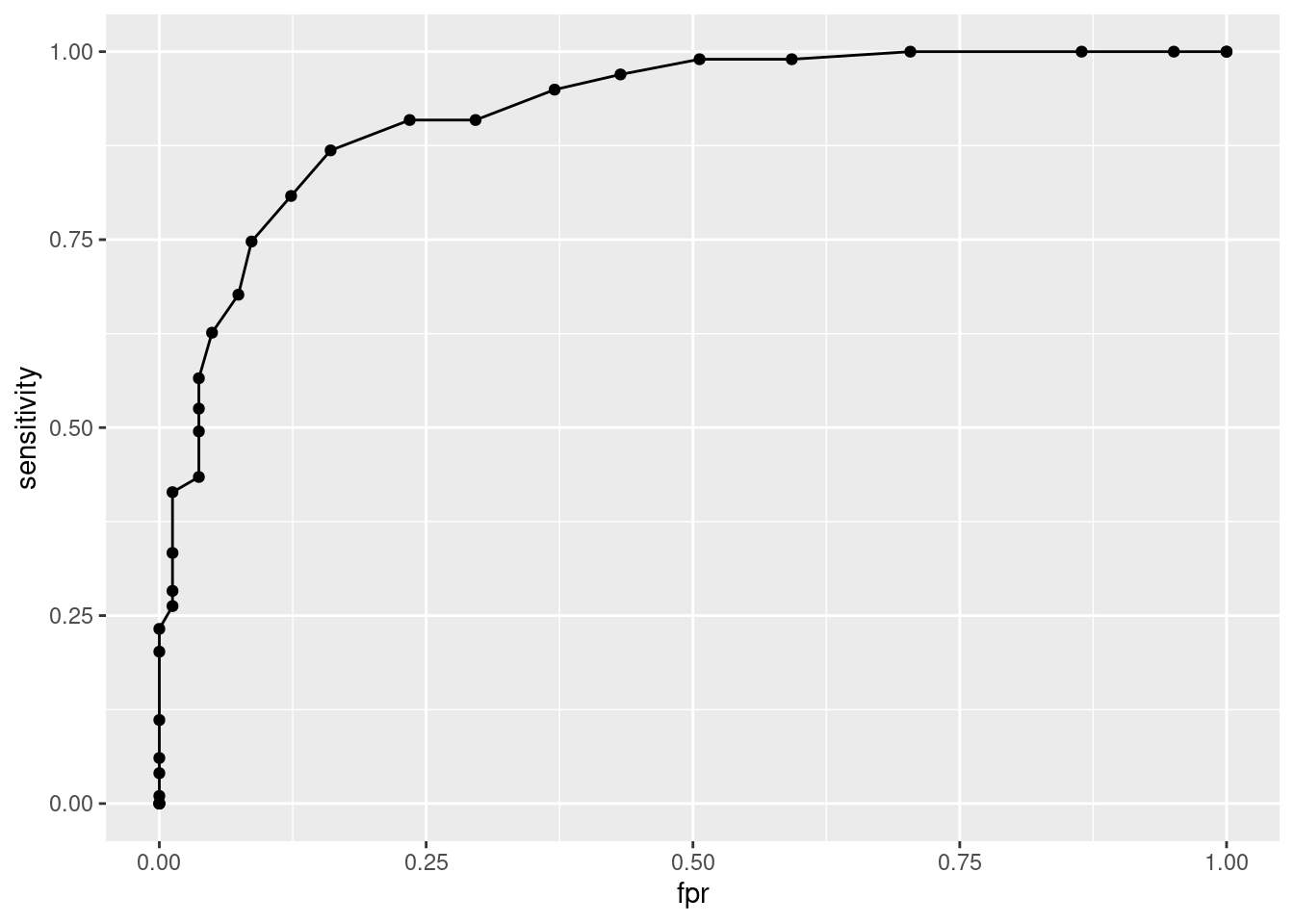
# AUC
dfpr <- c(diff(1 - specificity), 0)
dsens <- c(diff(sensitivity), 0)
sum(sensitivity * dfpr) + sum(dsens * dfpr) / 2[1] 0.920626You can also use the ROCit package
roc_adec <- rocit(dat1_sub$ADEC, class = dat1_sub$asd == "ASD")Warning in rocit(dat1_sub$ADEC, class = dat1_sub$asd == "ASD"): NA(s) in score
and/or class, removed from the data.plot(roc_adec)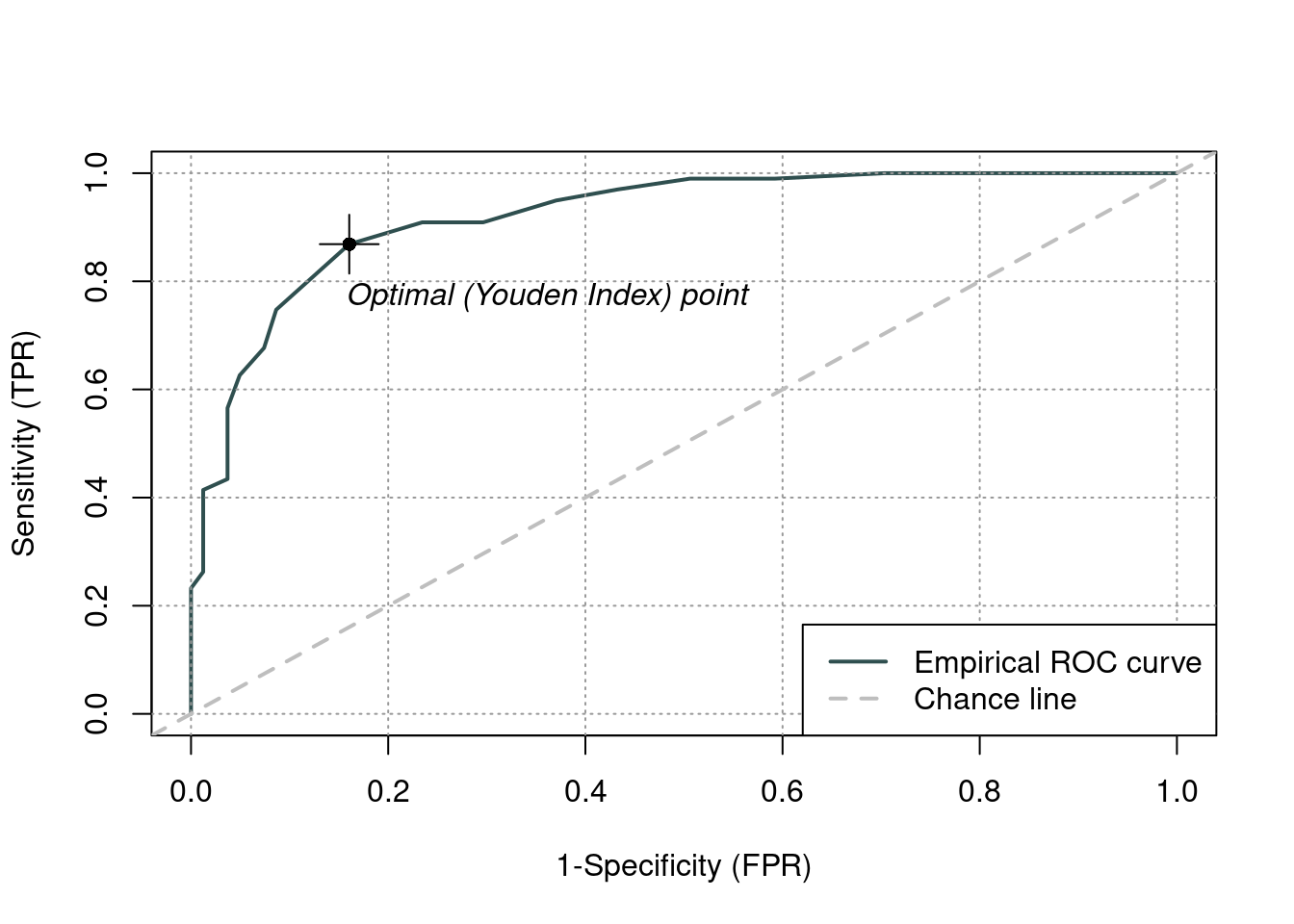
summary(roc_adec)
Method used: empirical
Number of positive(s): 99
Number of negative(s): 81
Area under curve: 0.9206 ciAUC(roc_adec)
estimated AUC : 0.920626013218606
AUC estimation method : empirical
CI of AUC
confidence level = 95%
lower = 0.880257283328194 upper = 0.960994743109017MTMM
Data from this paper: https://drive.google.com/file/d/1O4_MVWchC5kyCaf5p7GH4AjRIrY8zHJ8/
dat <- haven::read_sav("https://osf.io/download/5ayjb/")
dat_red <- haven::read_sav("https://osf.io/download/3uecq/")The data set contains scale scores for Neuroticism (N), Extraversion (E), Openness (O), Agreeableness (A), and Conscientiousness (C) with four methods of assessment: Self-reports (SR), from a close-other who is a female (RF), from a close-other who is a male (RM), implicit association tests (IAT), and behavioral assessment (BIH). You can see the labels in the SPSS data set using
# Loop over each column, and call `attr(which = "label")` for each column
# to obtain the label stored as an attribute
dat_labels <- lapply(dat_red, FUN = attr, which = "label")
# Put things into a table (tibble format)
tibble(variable = names(dat_red), label = unlist(dat_labels))To make the size of the matrix a bit more manageable, we will select only E, A, and C for illustration, and with the SR, RF, and IAT methods.
Descriptive Statistics
Here are the descriptives:
var_names <- c("ESR", "ASR", "CSR", "ERF", "ARF", "CRF", "EIAT", "AIAT", "CIAT")
datasummary_skim(dat_red[var_names])| Unique (#) | Missing (%) | Mean | SD | Min | Median | Max | ||
|---|---|---|---|---|---|---|---|---|
| ESR | 73 | 0 | 111.5 | 23.1 | 41.0 | 112.0 | 163.0 | |
| ASR | 79 | 0 | 114.9 | 24.3 | 35.0 | 116.5 | 163.0 | |
| CSR | 73 | 0 | 126.7 | 24.2 | 60.0 | 131.5 | 175.0 | |
| ERF | 63 | 0 | 111.7 | 21.0 | 47.0 | 112.0 | 159.0 | |
| ARF | 69 | 0 | 122.1 | 20.6 | 57.0 | 124.0 | 162.0 | |
| CRF | 67 | 0 | 140.7 | 22.5 | 75.0 | 141.0 | 182.0 | |
| EIAT | 146 | 0 | 0.2 | 0.4 | −1.1 | 0.2 | 1.1 | |
| AIAT | 146 | 0 | 0.5 | 0.4 | −0.5 | 0.5 | 1.3 | |
| CIAT | 146 | 0 | 0.5 | 0.4 | −1.0 | 0.5 | 1.6 |
MTMM Matrix
datasummary_correlation(dat_red[var_names])| ESR | ASR | CSR | ERF | ARF | CRF | EIAT | AIAT | CIAT | |
|---|---|---|---|---|---|---|---|---|---|
| ESR | 1 | . | . | . | . | . | . | . | . |
| ASR | .19 | 1 | . | . | . | . | . | . | . |
| CSR | .20 | .31 | 1 | . | . | . | . | . | . |
| ERF | .60 | .26 | .08 | 1 | . | . | . | . | . |
| ARF | .08 | .57 | .09 | .24 | 1 | . | . | . | . |
| CRF | .06 | .17 | .46 | .28 | .30 | 1 | . | . | . |
| EIAT | .07 | −.06 | .10 | .01 | −.03 | −.01 | 1 | . | . |
| AIAT | −.03 | −.12 | −.05 | −.19 | −.03 | .02 | .11 | 1 | . |
| CIAT | .01 | .03 | .08 | −.12 | .01 | .06 | .05 | .28 | 1 |