Item Response Theory
library(lme4)
library(mirt)Rasch Model
data(LSAT7, package = "mirt")
dat <- expand.table(LSAT7)m_rasch <- mirt(dat, model = 1, itemtype = "Rasch")
Iteration: 1, Log-Lik: -2664.942, Max-Change: 0.01075
Iteration: 2, Log-Lik: -2664.913, Max-Change: 0.00257
Iteration: 3, Log-Lik: -2664.910, Max-Change: 0.00210
Iteration: 4, Log-Lik: -2664.908, Max-Change: 0.00179
Iteration: 5, Log-Lik: -2664.906, Max-Change: 0.00156
Iteration: 6, Log-Lik: -2664.905, Max-Change: 0.00137
Iteration: 7, Log-Lik: -2664.904, Max-Change: 0.00141
Iteration: 8, Log-Lik: -2664.903, Max-Change: 0.00110
Iteration: 9, Log-Lik: -2664.903, Max-Change: 0.00095
Iteration: 10, Log-Lik: -2664.902, Max-Change: 0.00086
Iteration: 11, Log-Lik: -2664.902, Max-Change: 0.00072
Iteration: 12, Log-Lik: -2664.902, Max-Change: 0.00063
Iteration: 13, Log-Lik: -2664.902, Max-Change: 0.00065
Iteration: 14, Log-Lik: -2664.901, Max-Change: 0.00051
Iteration: 15, Log-Lik: -2664.901, Max-Change: 0.00044
Iteration: 16, Log-Lik: -2664.901, Max-Change: 0.00040
Iteration: 17, Log-Lik: -2664.901, Max-Change: 0.00034
Iteration: 18, Log-Lik: -2664.901, Max-Change: 0.00029
Iteration: 19, Log-Lik: -2664.901, Max-Change: 0.00030
Iteration: 20, Log-Lik: -2664.901, Max-Change: 0.00024
Iteration: 21, Log-Lik: -2664.901, Max-Change: 0.00020
Iteration: 22, Log-Lik: -2664.901, Max-Change: 0.00018
Iteration: 23, Log-Lik: -2664.901, Max-Change: 0.00016
Iteration: 24, Log-Lik: -2664.901, Max-Change: 0.00014
Iteration: 25, Log-Lik: -2664.901, Max-Change: 0.00014
Iteration: 26, Log-Lik: -2664.901, Max-Change: 0.00011
Iteration: 27, Log-Lik: -2664.901, Max-Change: 0.00009coef(m_rasch, IRTpars = TRUE) # d = intercept = -1 * difficulty$Item.1
a b g u
par 1 -1.868 0 1
$Item.2
a b g u
par 1 -0.791 0 1
$Item.3
a b g u
par 1 -1.461 0 1
$Item.4
a b g u
par 1 -0.521 0 1
$Item.5
a b g u
par 1 -1.993 0 1
$GroupPars
MEAN_1 COV_11
par 0 1.022The 1PL is just a reparameterization of the Rasch model:
# 1PL
mod_1pl <- "
THETA = 1-5
CONSTRAIN = (1-5, a1)
"
m_1pl <- mirt(dat, mod_1pl)
Iteration: 1, Log-Lik: -2668.786, Max-Change: 0.06198
Iteration: 2, Log-Lik: -2666.196, Max-Change: 0.03935
Iteration: 3, Log-Lik: -2665.347, Max-Change: 0.02405
Iteration: 4, Log-Lik: -2664.955, Max-Change: 0.00867
Iteration: 5, Log-Lik: -2664.920, Max-Change: 0.00509
Iteration: 6, Log-Lik: -2664.907, Max-Change: 0.00294
Iteration: 7, Log-Lik: -2664.902, Max-Change: 0.00135
Iteration: 8, Log-Lik: -2664.901, Max-Change: 0.00079
Iteration: 9, Log-Lik: -2664.901, Max-Change: 0.00046
Iteration: 10, Log-Lik: -2664.901, Max-Change: 0.00022
Iteration: 11, Log-Lik: -2664.901, Max-Change: 0.00011
Iteration: 12, Log-Lik: -2664.901, Max-Change: 0.00006coef(m_1pl, IRTpars = TRUE) # b = difficulty$Item.1
a b g u
par 1.011 -1.848 0 1
$Item.2
a b g u
par 1.011 -0.782 0 1
$Item.3
a b g u
par 1.011 -1.445 0 1
$Item.4
a b g u
par 1.011 -0.516 0 1
$Item.5
a b g u
par 1.011 -1.971 0 1
$GroupPars
MEAN_1 COV_11
par 0 1Standard Errors
m_rasch <- mirt(dat, model = 1, itemtype = "Rasch", SE = TRUE)
Iteration: 1, Log-Lik: -2664.942, Max-Change: 0.01075
Iteration: 2, Log-Lik: -2664.913, Max-Change: 0.00257
Iteration: 3, Log-Lik: -2664.910, Max-Change: 0.00210
Iteration: 4, Log-Lik: -2664.908, Max-Change: 0.00179
Iteration: 5, Log-Lik: -2664.906, Max-Change: 0.00156
Iteration: 6, Log-Lik: -2664.905, Max-Change: 0.00137
Iteration: 7, Log-Lik: -2664.904, Max-Change: 0.00141
Iteration: 8, Log-Lik: -2664.903, Max-Change: 0.00110
Iteration: 9, Log-Lik: -2664.903, Max-Change: 0.00095
Iteration: 10, Log-Lik: -2664.902, Max-Change: 0.00086
Iteration: 11, Log-Lik: -2664.902, Max-Change: 0.00072
Iteration: 12, Log-Lik: -2664.902, Max-Change: 0.00063
Iteration: 13, Log-Lik: -2664.902, Max-Change: 0.00065
Iteration: 14, Log-Lik: -2664.901, Max-Change: 0.00051
Iteration: 15, Log-Lik: -2664.901, Max-Change: 0.00044
Iteration: 16, Log-Lik: -2664.901, Max-Change: 0.00040
Iteration: 17, Log-Lik: -2664.901, Max-Change: 0.00034
Iteration: 18, Log-Lik: -2664.901, Max-Change: 0.00029
Iteration: 19, Log-Lik: -2664.901, Max-Change: 0.00030
Iteration: 20, Log-Lik: -2664.901, Max-Change: 0.00024
Iteration: 21, Log-Lik: -2664.901, Max-Change: 0.00020
Iteration: 22, Log-Lik: -2664.901, Max-Change: 0.00018
Iteration: 23, Log-Lik: -2664.901, Max-Change: 0.00016
Iteration: 24, Log-Lik: -2664.901, Max-Change: 0.00014
Iteration: 25, Log-Lik: -2664.901, Max-Change: 0.00014
Iteration: 26, Log-Lik: -2664.901, Max-Change: 0.00011
Iteration: 27, Log-Lik: -2664.901, Max-Change: 0.00009
Calculating information matrix...coef(m_rasch, IRTpars = TRUE)$Item.1
a b g u
par 1 -1.868 0 1
CI_2.5 NA -2.065 NA NA
CI_97.5 NA -1.671 NA NA
$Item.2
a b g u
par 1 -0.791 0 1
CI_2.5 NA -0.950 NA NA
CI_97.5 NA -0.632 NA NA
$Item.3
a b g u
par 1 -1.461 0 1
CI_2.5 NA -1.640 NA NA
CI_97.5 NA -1.282 NA NA
$Item.4
a b g u
par 1 -0.521 0 1
CI_2.5 NA -0.676 NA NA
CI_97.5 NA -0.367 NA NA
$Item.5
a b g u
par 1 -1.993 0 1
CI_2.5 NA -2.196 NA NA
CI_97.5 NA -1.790 NA NA
$GroupPars
MEAN_1 COV_11
par 0 1.022
CI_2.5 NA 0.765
CI_97.5 NA 1.279Plots
Item Response Function
Also item characteristic curve
plot(m_rasch, type = "trace")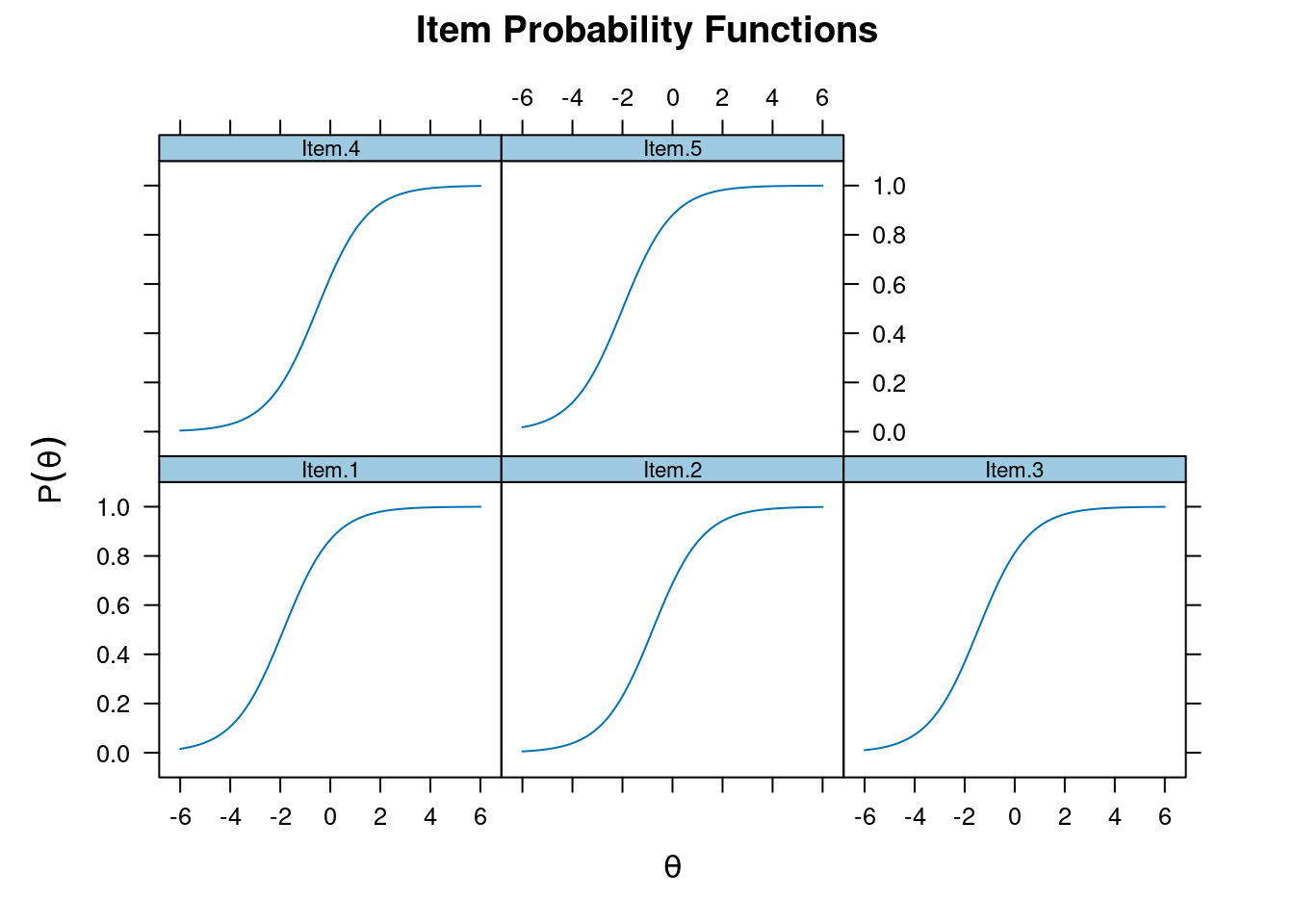
Item and Test Information
Item information
plot(m_rasch, type = "infotrace")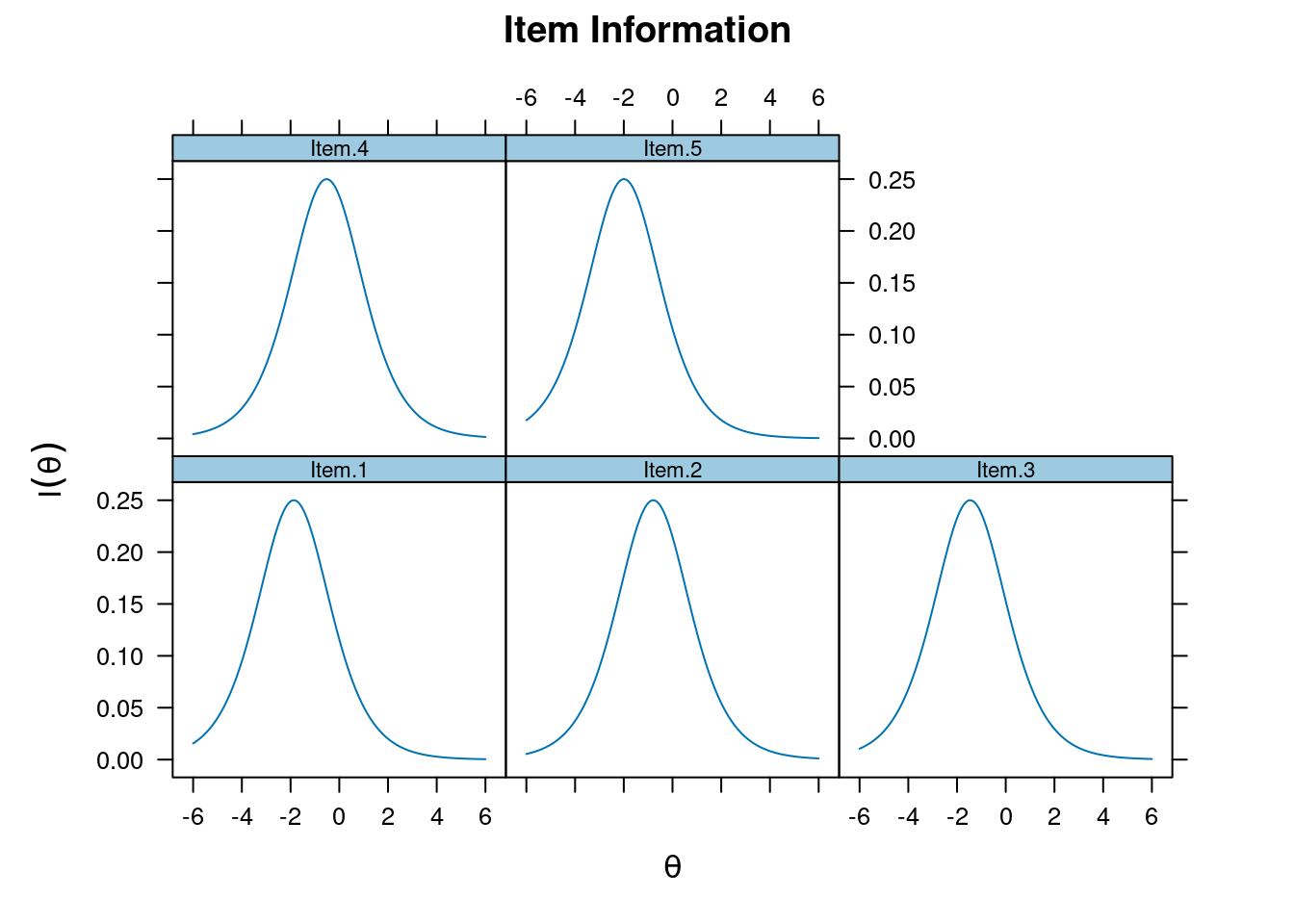
plot(m_rasch, type = "info")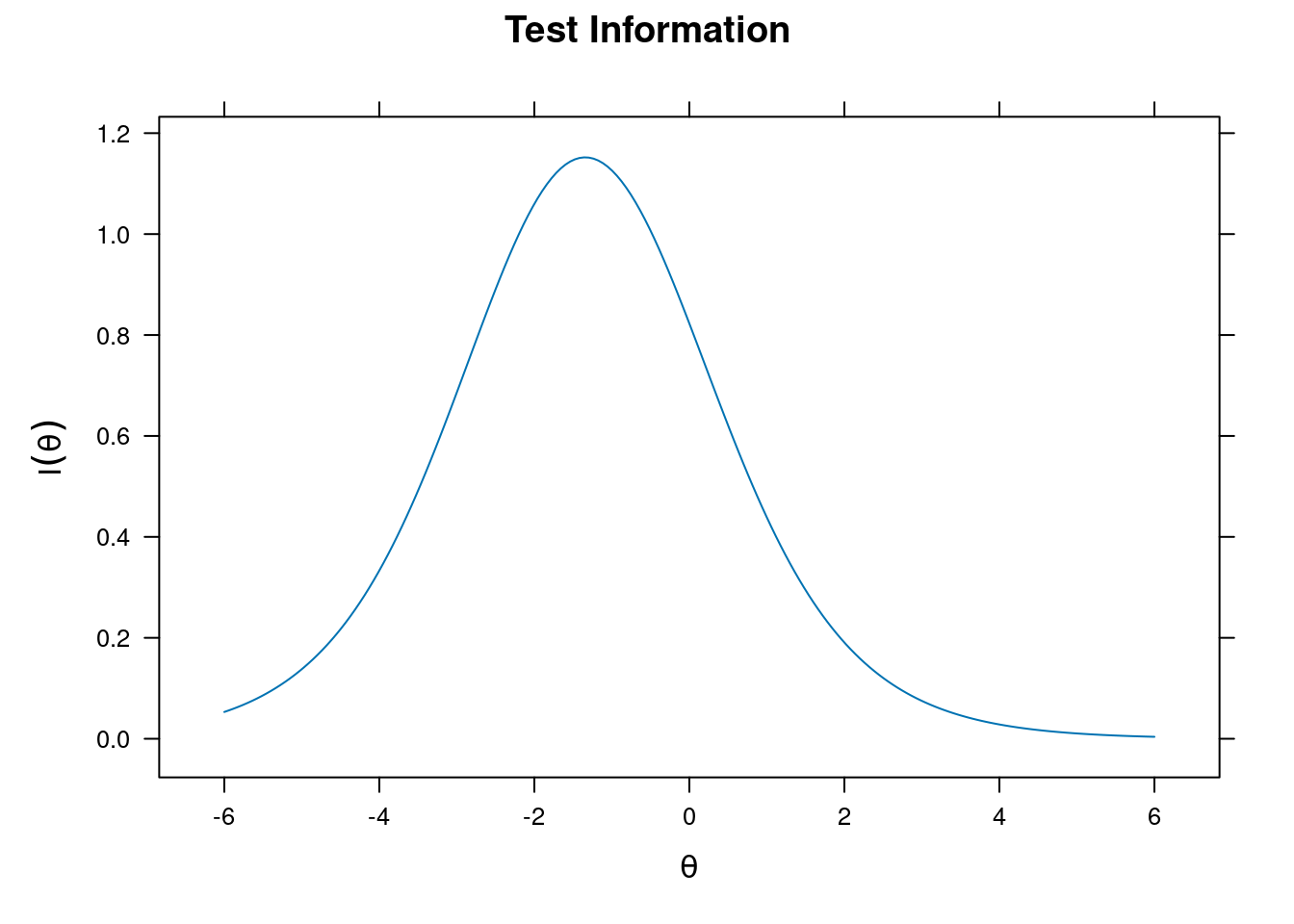
Assumptions/Model Fit
Global test statistics (analogous to those in factor analysis)
- \(G^2\) test statistic (not valid when matrix is sparse)
- \(M_2\): better approximation for polytomous models
m_rasch
Call:
mirt(data = dat, model = 1, itemtype = "Rasch", SE = TRUE)
Full-information item factor analysis with 1 factor(s).
Converged within 1e-04 tolerance after 27 EM iterations.
mirt version: 1.38.1
M-step optimizer: nlminb
EM acceleration: Ramsay
Number of rectangular quadrature: 61
Latent density type: Gaussian
Information matrix estimated with method: Oakes
Second-order test: model is a possible local maximum
Condition number of information matrix = 4.488772
Log-likelihood = -2664.901
Estimated parameters: 6
AIC = 5341.802
BIC = 5371.248; SABIC = 5352.192
G2 (25) = 43.89, p = 0.0112
RMSEA = 0.028, CFI = NaN, TLI = NaNM2(m_rasch)Parallel analysis
psych::fa.parallel(dat, cor = "poly")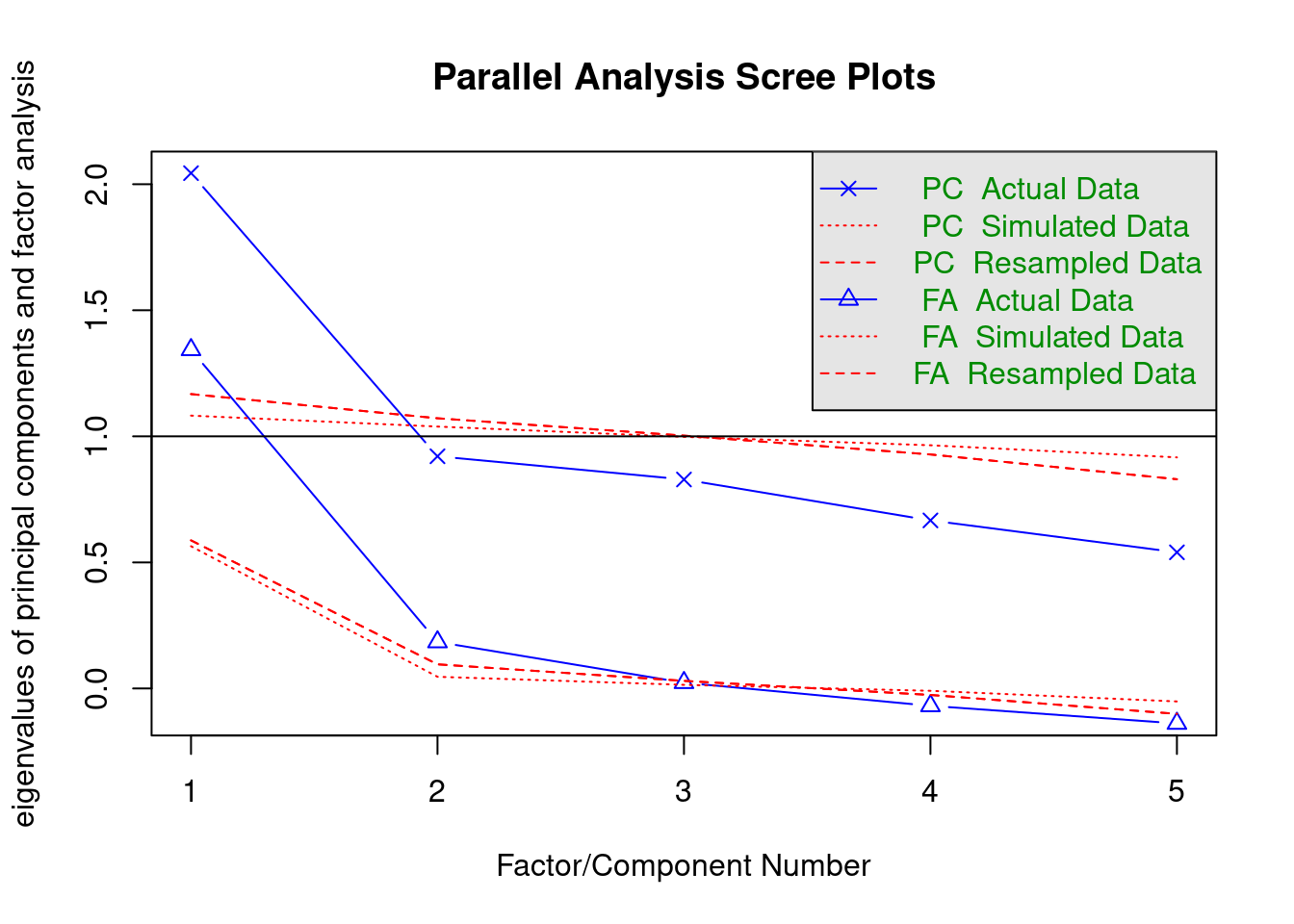
Parallel analysis suggests that the number of factors = 2 and the number of components = 1 DIMTEST
# Need the sirt package. See the conf.detect() function
# Need to obtain WLE scores first
wle_scores <- fscores(m_rasch, method = "WLE")
detect1 <- sirt::conf.detect(dat,
score = as.vector(wle_scores),
itemcluster = rep(1, 5)
)-----------------------------------------------------------
Confirmatory DETECT Analysis
Conditioning on 1 Score
Bandwidth Scale: 1.1
Pairwise Estimation of Conditional Covariances
...........................................................
Nonparametric ICC estimation
...........................................................
Nonparametric Estimation of conditional covariances
5% 10% 15% 20% 25% 30% 35% 40% 45% 50%
-----------------------------------------------------------
unweighted weighted
DETECT -3.437 -3.437
ASSI -1.000 -1.000
RATIO -1.000 -1.000
MADCOV100 3.437 3.437
MCOV100 -3.437 -3.437# DETECT < .20 suggest essential unidimensionalityLocal Independence
residuals(m_rasch, type = "Q3") # Q3Q3 summary statistics:
Min. 1st Qu. Median Mean 3rd Qu. Max.
-0.196 -0.145 -0.116 -0.110 -0.082 0.000
Item.1 Item.2 Item.3 Item.4 Item.5
Item.1 1.000 -0.136 -0.099 -0.080 -0.051
Item.2 -0.136 1.000 0.000 -0.196 -0.169
Item.3 -0.099 0.000 1.000 -0.133 -0.090
Item.4 -0.080 -0.196 -0.133 1.000 -0.149
Item.5 -0.051 -0.169 -0.090 -0.149 1.000# All |Q3| < .2 in this caseItem Fit
S-\(\chi^2\)
itemfit(m_rasch)# For large number of items, the p values should be adjusted
itemfit(m_rasch, p.adjust = "holm")Two-Parameter Logistic
m_2pl <- mirt(dat, model = 1, itemtype = "2PL")
Iteration: 1, Log-Lik: -2668.786, Max-Change: 0.18243
Iteration: 2, Log-Lik: -2663.691, Max-Change: 0.13637
Iteration: 3, Log-Lik: -2661.454, Max-Change: 0.10231
Iteration: 4, Log-Lik: -2659.430, Max-Change: 0.04181
Iteration: 5, Log-Lik: -2659.241, Max-Change: 0.03417
Iteration: 6, Log-Lik: -2659.113, Max-Change: 0.02911
Iteration: 7, Log-Lik: -2658.812, Max-Change: 0.00456
Iteration: 8, Log-Lik: -2658.809, Max-Change: 0.00363
Iteration: 9, Log-Lik: -2658.808, Max-Change: 0.00273
Iteration: 10, Log-Lik: -2658.806, Max-Change: 0.00144
Iteration: 11, Log-Lik: -2658.806, Max-Change: 0.00118
Iteration: 12, Log-Lik: -2658.806, Max-Change: 0.00101
Iteration: 13, Log-Lik: -2658.805, Max-Change: 0.00042
Iteration: 14, Log-Lik: -2658.805, Max-Change: 0.00025
Iteration: 15, Log-Lik: -2658.805, Max-Change: 0.00026
Iteration: 16, Log-Lik: -2658.805, Max-Change: 0.00023
Iteration: 17, Log-Lik: -2658.805, Max-Change: 0.00023
Iteration: 18, Log-Lik: -2658.805, Max-Change: 0.00021
Iteration: 19, Log-Lik: -2658.805, Max-Change: 0.00019
Iteration: 20, Log-Lik: -2658.805, Max-Change: 0.00017
Iteration: 21, Log-Lik: -2658.805, Max-Change: 0.00017
Iteration: 22, Log-Lik: -2658.805, Max-Change: 0.00015
Iteration: 23, Log-Lik: -2658.805, Max-Change: 0.00015
Iteration: 24, Log-Lik: -2658.805, Max-Change: 0.00013
Iteration: 25, Log-Lik: -2658.805, Max-Change: 0.00013
Iteration: 26, Log-Lik: -2658.805, Max-Change: 0.00011
Iteration: 27, Log-Lik: -2658.805, Max-Change: 0.00011
Iteration: 28, Log-Lik: -2658.805, Max-Change: 0.00010coef(m_2pl, IRTpars = TRUE)$Item.1
a b g u
par 0.988 -1.879 0 1
$Item.2
a b g u
par 1.081 -0.748 0 1
$Item.3
a b g u
par 1.706 -1.058 0 1
$Item.4
a b g u
par 0.765 -0.635 0 1
$Item.5
a b g u
par 0.736 -2.52 0 1
$GroupPars
MEAN_1 COV_11
par 0 1plot(m_rasch, type = "trace")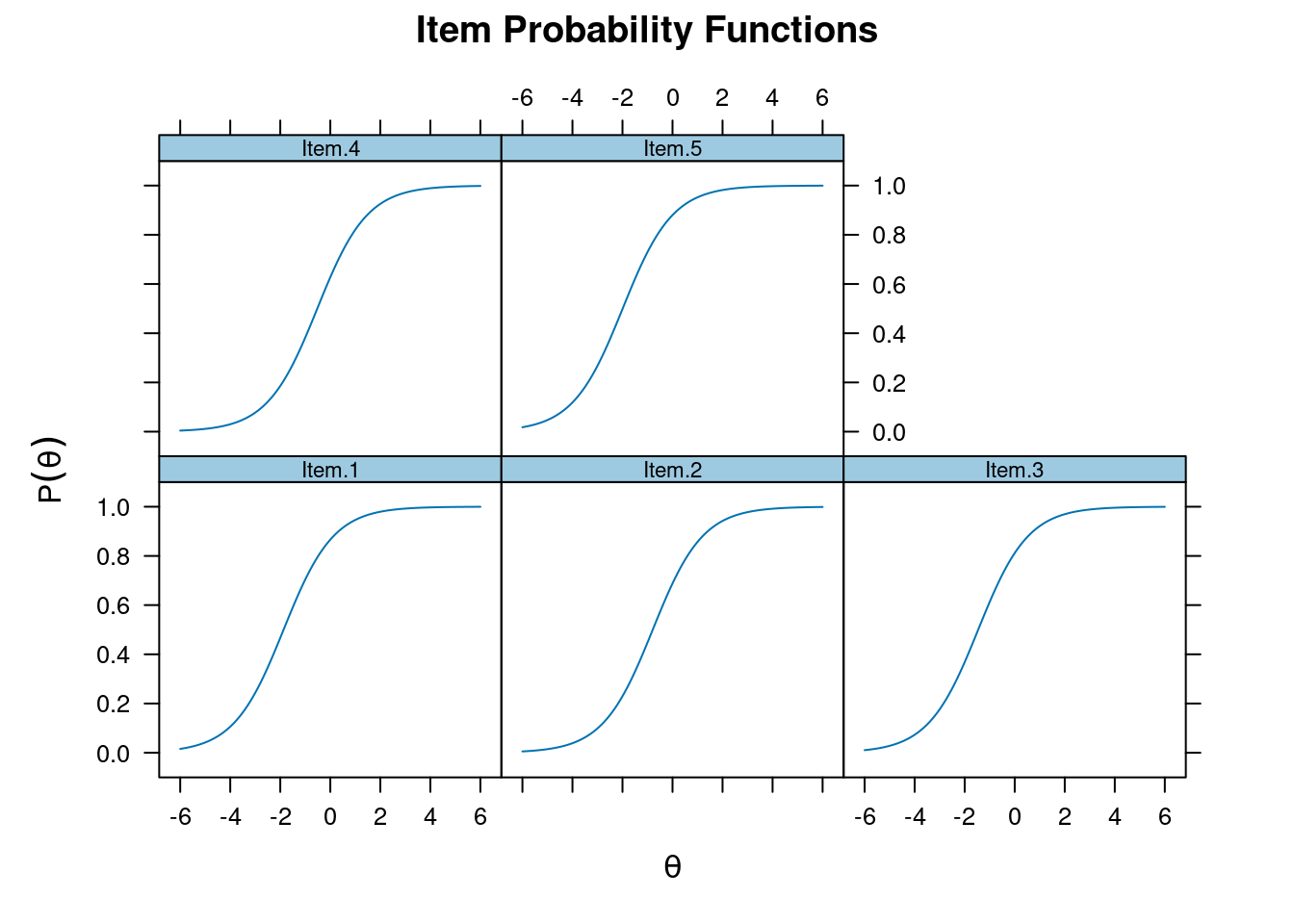
Response function on the same canvas
plot(m_2pl, type = "trace", facet_items = FALSE)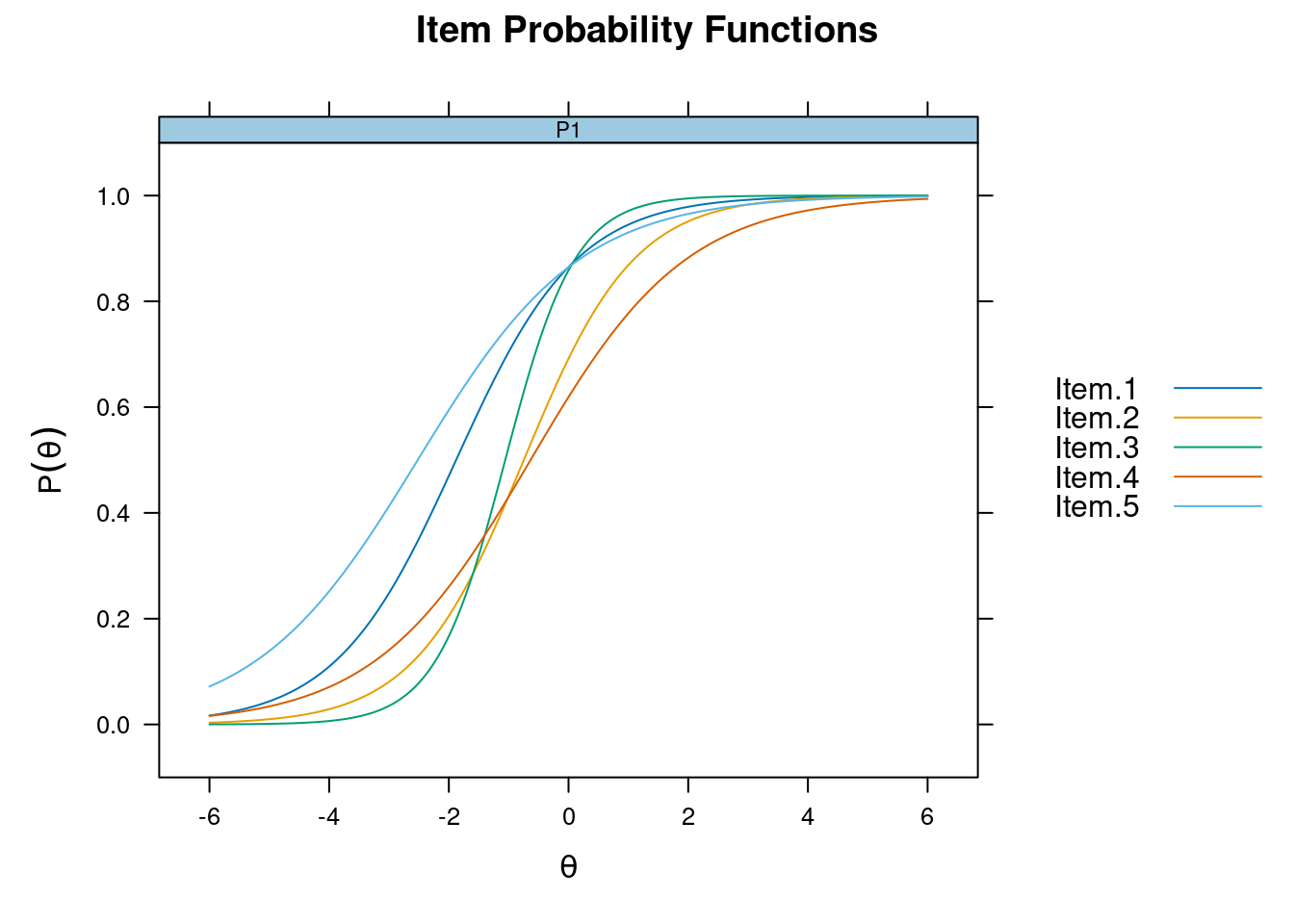
Model Comparison
anova(m_rasch, m_2pl)Some Polytomous Models
Rating scale model
data(bfi, package = "psych")
# Neuroticism
m_rsm <- mirt(bfi[16:20], itemtype = "rsm")
Iteration: 1, Log-Lik: -23985.274, Max-Change: 1.00910
Iteration: 2, Log-Lik: -22434.284, Max-Change: 0.18792
Iteration: 3, Log-Lik: -22338.797, Max-Change: 0.17826
Iteration: 4, Log-Lik: -22282.201, Max-Change: 0.12715
Iteration: 5, Log-Lik: -22244.160, Max-Change: 0.09083
Iteration: 6, Log-Lik: -22217.562, Max-Change: 0.06646
Iteration: 7, Log-Lik: -22198.665, Max-Change: 0.04953
Iteration: 8, Log-Lik: -22185.225, Max-Change: 0.03738
Iteration: 9, Log-Lik: -22175.730, Max-Change: 0.02839
Iteration: 10, Log-Lik: -22169.095, Max-Change: 0.02270
Iteration: 11, Log-Lik: -22164.507, Max-Change: 0.01858
Iteration: 12, Log-Lik: -22161.370, Max-Change: 0.01513
Iteration: 13, Log-Lik: -22156.185, Max-Change: 0.01992
Iteration: 14, Log-Lik: -22155.234, Max-Change: 0.00485
Iteration: 15, Log-Lik: -22155.115, Max-Change: 0.00311
Iteration: 16, Log-Lik: -22154.996, Max-Change: 0.00344
Iteration: 17, Log-Lik: -22154.963, Max-Change: 0.00141
Iteration: 18, Log-Lik: -22154.950, Max-Change: 0.00099
Iteration: 19, Log-Lik: -22154.935, Max-Change: 0.00132
Iteration: 20, Log-Lik: -22154.930, Max-Change: 0.00040
Iteration: 21, Log-Lik: -22154.929, Max-Change: 0.00027
Iteration: 22, Log-Lik: -22154.928, Max-Change: 0.00033
Iteration: 23, Log-Lik: -22154.928, Max-Change: 0.00012
Iteration: 24, Log-Lik: -22154.928, Max-Change: 0.00008itemfit(m_rsm, na.rm = TRUE) # not fitting wellSample size after row-wise response data removal: 2694plot(m_rsm, type = "trace")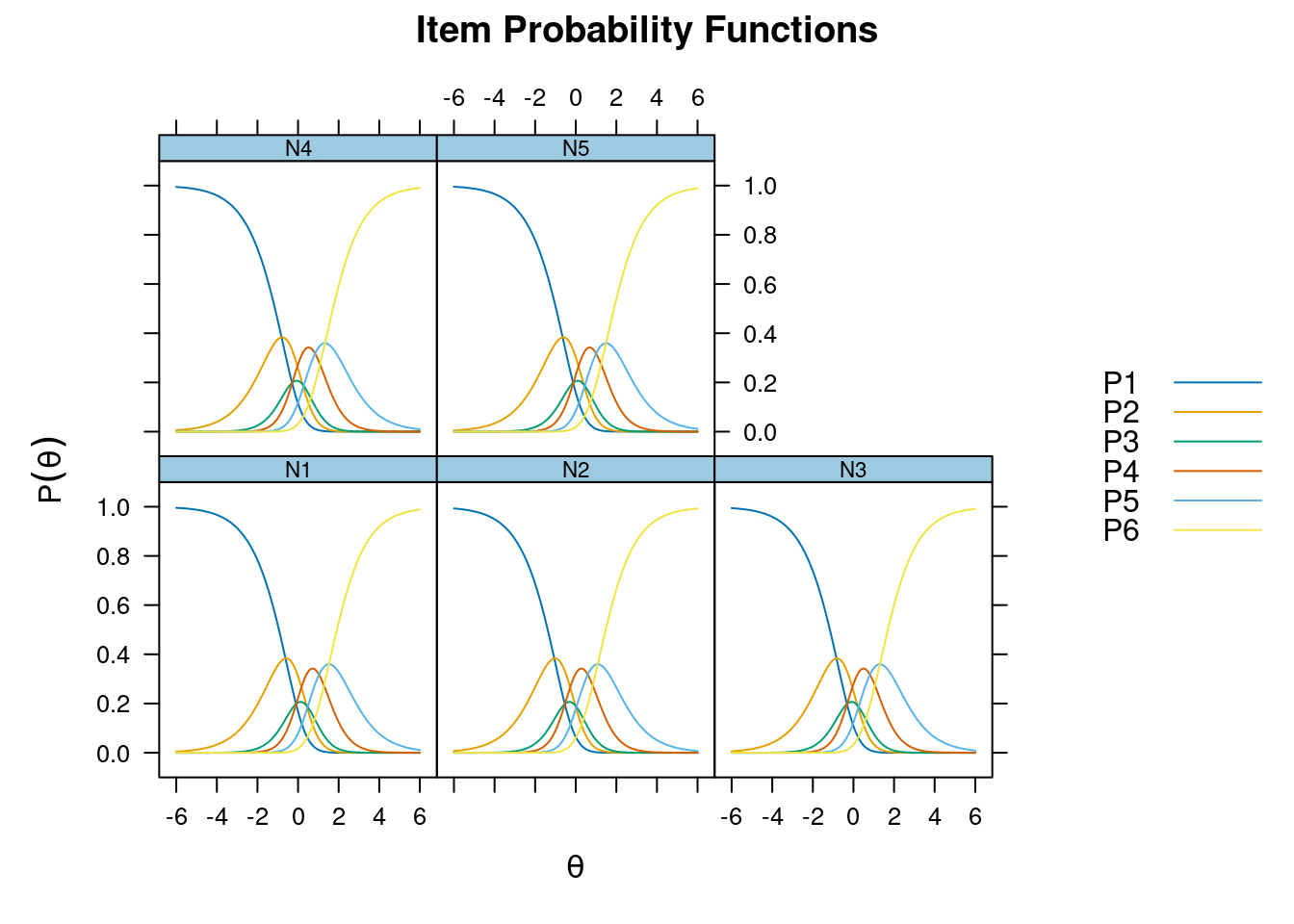
Graded Response model
m_grm <- mirt(bfi[16:20], itemtype = "graded")
Iteration: 1, Log-Lik: -22678.849, Max-Change: 0.98180
Iteration: 2, Log-Lik: -22009.516, Max-Change: 0.62057
Iteration: 3, Log-Lik: -21843.393, Max-Change: 0.31998
Iteration: 4, Log-Lik: -21773.416, Max-Change: 0.25006
Iteration: 5, Log-Lik: -21747.311, Max-Change: 0.16264
Iteration: 6, Log-Lik: -21734.951, Max-Change: 0.15148
Iteration: 7, Log-Lik: -21728.377, Max-Change: 0.09627
Iteration: 8, Log-Lik: -21724.841, Max-Change: 0.07974
Iteration: 9, Log-Lik: -21722.917, Max-Change: 0.06180
Iteration: 10, Log-Lik: -21721.933, Max-Change: 0.02070
Iteration: 11, Log-Lik: -21721.615, Max-Change: 0.01547
Iteration: 12, Log-Lik: -21721.511, Max-Change: 0.01199
Iteration: 13, Log-Lik: -21721.392, Max-Change: 0.00246
Iteration: 14, Log-Lik: -21721.388, Max-Change: 0.00214
Iteration: 15, Log-Lik: -21721.385, Max-Change: 0.00159
Iteration: 16, Log-Lik: -21721.382, Max-Change: 0.00202
Iteration: 17, Log-Lik: -21721.381, Max-Change: 0.00050
Iteration: 18, Log-Lik: -21721.381, Max-Change: 0.00008itemfit(m_grm, na.rm = TRUE) # not fitting wellSample size after row-wise response data removal: 2694plot(m_grm, type = "trace")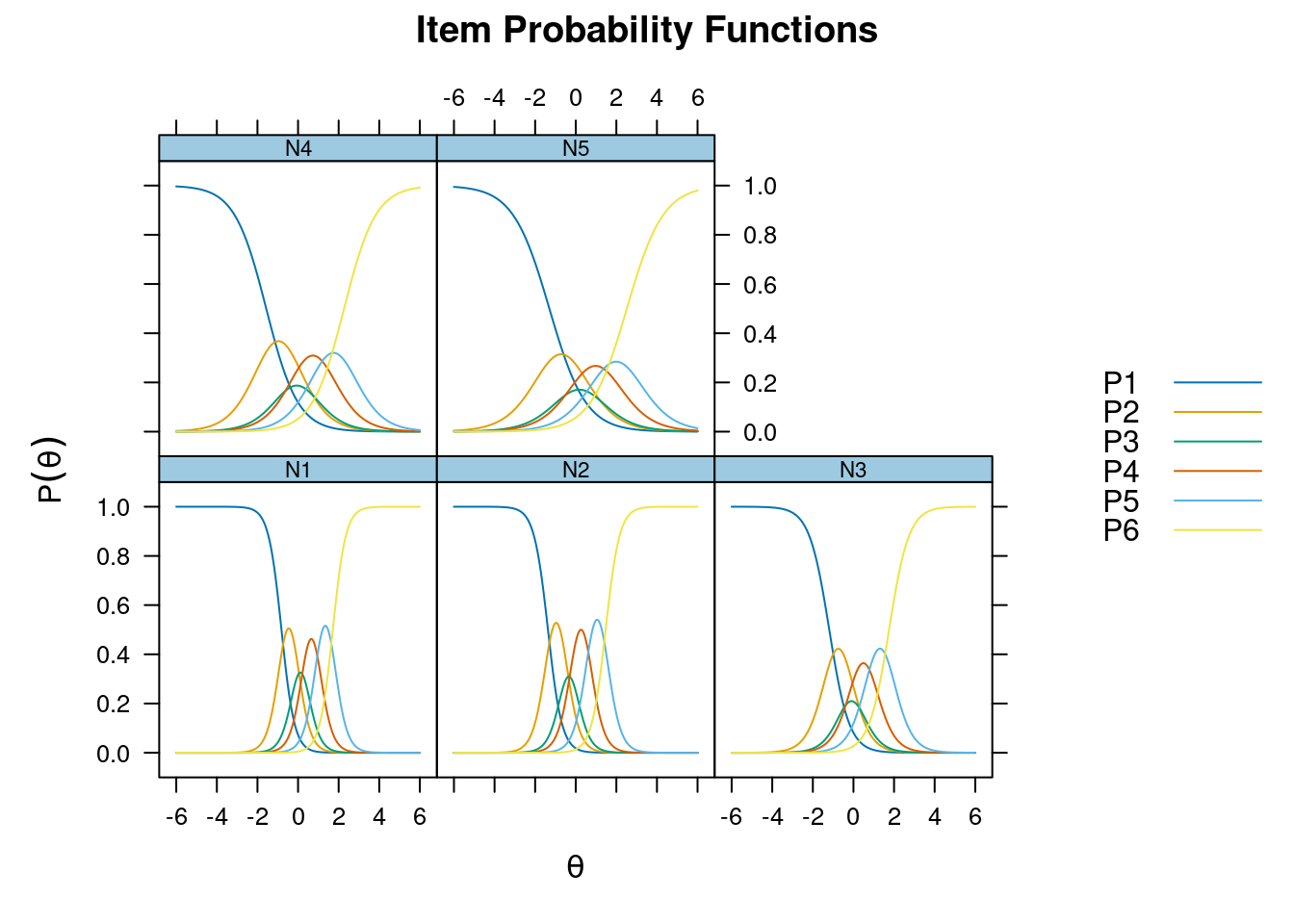
plot(m_grm, type = "info")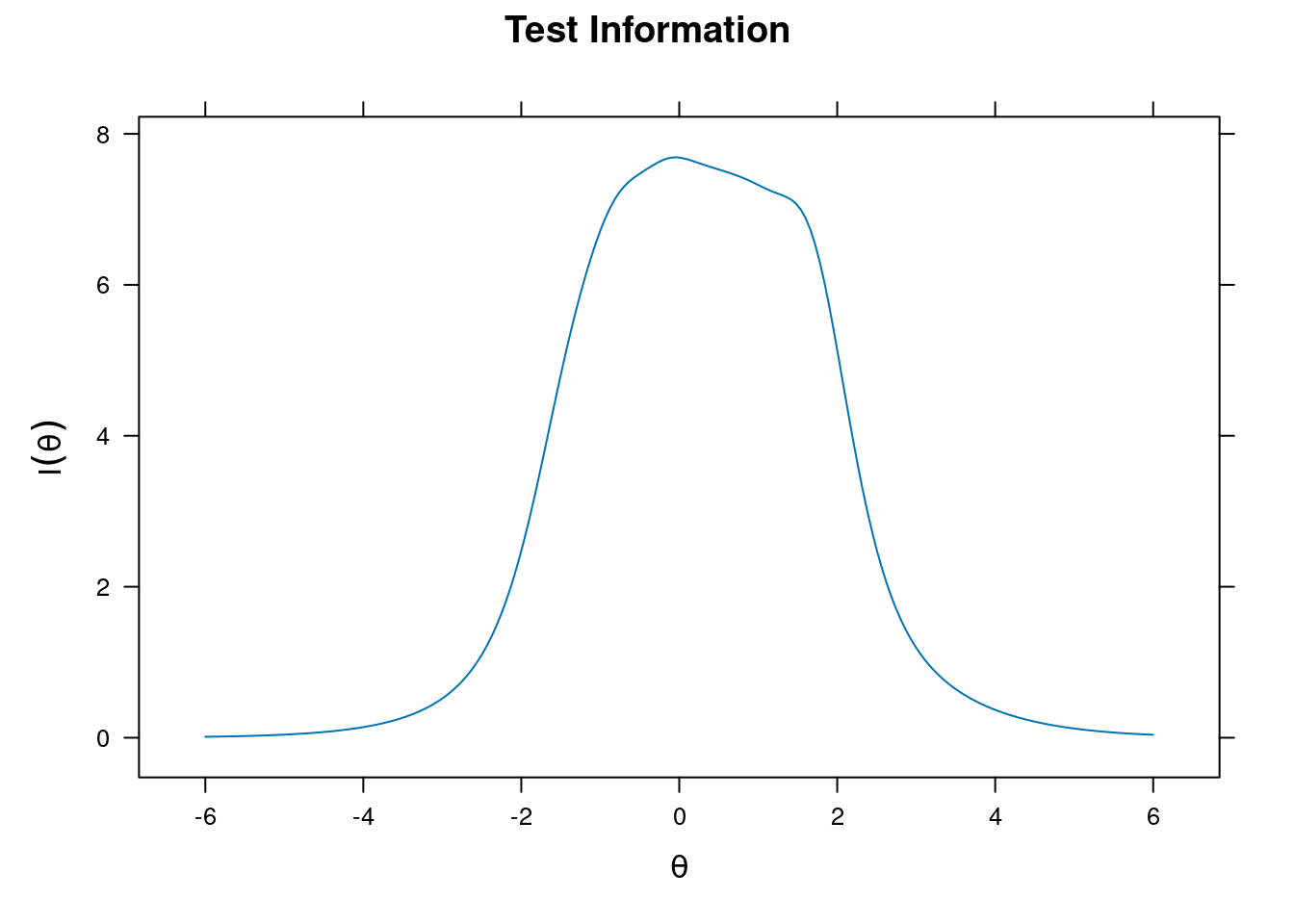
Estimation of 1-PL Model
For simplicity, I assume \(Da = 1\) here; in practice \(a\) is usually estimated with 1-PL.
Conditional likelihood (assumed \(\theta\) known)
Let \(P_i(\theta_j)\) = \(P(Y_{i} = 1 \mid \theta = \theta_j)\)
If \(Y_{ij} = 1\),
\[\mathcal{L}(b_i; y_{ij}, \theta_j) = P_i(\theta_j)\]
If \(y_{ij} = 0\),
\[\mathcal{L}(b_i; y_{ij}, \theta_j) = 1 - P_i(\theta_j)\]
Marginal Likelihood (Integrating out \(\theta\))
Let \(\theta_j \sim N(0, 1)\), meaning its probability density is
\[f(\theta) = \Phi(\theta) = \frac{1}{\sqrt{2 \pi} \sigma} e^{-\theta^2 / 2}\]
\[\mathcal{L}(b_i; Y_{ij} = 1) = \int_{-\infty}^\infty P_i(t) \Phi(t) \,\mathrm{d} t\]
and
\[\ell(b_i; Y_{ij} = 1) = \log \left[\int_{-\infty}^\infty P_i(t) \Phi(t) \,\mathrm{d} t\right]\]
Combining \(N\) independent observations, and let \(\bar y_i\) be the mean of item \(i\) (i.e., proportion scoring 1),
\[ \begin{aligned} \ell(b_i; y_{i1}, y_{i2}, \ldots, y_{iN}) & = N \left\{\bar y_i \log \left[\int_{-\infty}^\infty P_i(t) \Phi(t) \,\mathrm{d} t \right] \right. \\ & \quad \left. {} + (1 - \bar y_i) \log \left[1 - \int_{-\infty}^\infty P_i(t) \Phi(t) \,\mathrm{d} t \right]\right\} \end{aligned} \]
Maximum likelihood estimation finds values of \(b_i\) that maximizes the above quantity. However, the above quantity involves an integral, which does not generally have an analytic solution. More typically, we use Gaussian-Hermite quadrature to approximate the integral.
Quadrature
See https://en.wikipedia.org/wiki/Gauss%E2%80%93Hermite_quadrature
# Approximate z^2 for z ~ N(0, 1)
ghq5 <- lme4::GHrule(5)
sum(ghq5[, "w"] * ghq5[, "z"]^2)[1] 1pj <- function(theta, bj) plogis(theta - bj)
sum(ghq5[, "w"] * pj(ghq5[, "z"], bj = 1))[1] 0.303218sum(ghq5[, "w"] * pj(ghq5[, "z"], bj = 2))[1] 0.1555028loglik_1pl <- function(b, n, ybar, gh_ord = 5) {
ghq <- lme4::GHrule(gh_ord)
int_pj <- sum(ghq[, "w"] * pj(ghq[, "z"], bj = b))
n * (ybar * log(int_pj) + (1 - ybar) * log1p(-int_pj))
}
loglik_1pl(1, n = 1000, ybar = 828 / 1000)[1] -1050.196# Vectorized version
pj <- function(theta, bj) plogis(outer(theta, Y = bj, FUN = "-"))
loglik_1plj <- function(b, n, ybar, ghq = lme4::GHrule(5)) {
int_pj <- crossprod(ghq[, "w"], pj(ghq[, "z"], bj = b))
as.vector(
n * (ybar * log(int_pj) + (1 - ybar) * log1p(-int_pj))
)
}
loglik_1plj(c(-1, 0, 1), n = 1000, ybar = 828 / 1000)[1] -504.3902 -693.1472 -1050.1957curve(loglik_1plj(x, n = 1000, ybar = 828 /1000),
from = -4, to = 4,
xlab = expression(b[i]),
ylab = "log-likelihood")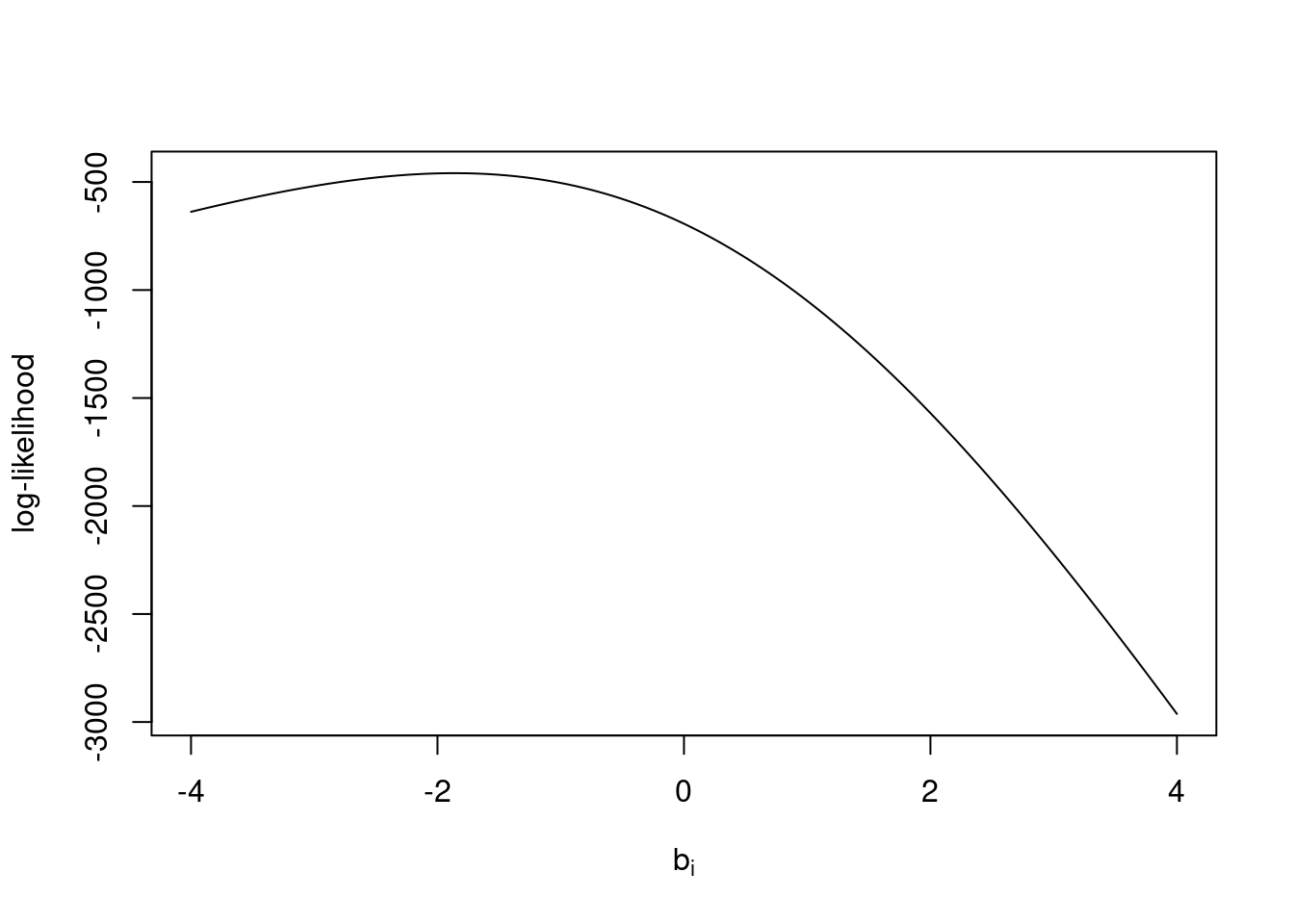
# Optimization
optim(0,
fn = loglik_1plj,
n = 1000, ybar = 828 / 1000,
method = "L-BFGS-B",
# Minimize -2 x ll
control = list(fnscale = -2)
)$par
[1] -1.862783
$value
[1] -459.0433
$counts
function gradient
7 7
$convergence
[1] 0
$message
[1] "CONVERGENCE: REL_REDUCTION_OF_F <= FACTR*EPSMCH"While here we assume \(a\) is known, in reality, \(a\) needs to be estimated as well and require we obtain the joint likelihood by adding up the likelihood across all items (assuming local independence).